FreeSurfer: recon-all-clinical#
This notebook was prepared for an audience of clinicians at the 2025 PACTALS conference (Pan-Asian consortium of Treatment and Research in ALS) in Melbourne.
Author: Thomas Shaw, 1 Sept 2025
Citations and Resources:#
Tools included in this workflow#
recon-all-clinical:
Karthik Gopinath, Douglas N. Greve, Colin Magdamo, Steve Arnold, Sudeshna Das, Oula Puonti, Juan Eugenio Iglesias, “Recon-all-clinical”: Cortical surface reconstruction and analysis of heterogeneous clinical brain MRI, Medical Image Analysis, Volume 103, 2025, 103608, ISSN 1361-8415, https://doi.org/10.1016/j.media.2025.103608.
Dataset:#
T1 weighted MP2RAGE at 7T (healthy control)
Shaw TB, York A, Barth M, Bollmann S. Towards Optimising MRI Characterisation of Tissue (TOMCAT) Dataset including all Longitudinal Automatic Segmentation of Hippocampal Subfields (LASHiS) data. Data Brief. 2020 Jul 20;32:106043. doi: https://doi.org/10.1016/j.dib.2020.106043 PMID: 32793772; PMCID: PMC7415822.
General Instructions:#
📘 How to Use This Notebook
This notebook is an interactive document. It mixes short explanations (like this box) with computer code that runs automatically.
⚡ The basics
- 👆 Click on a cell (the boxes with text or code)
- ▶️ Run it by pressing
Command + Enter(Mac) orControl + Enter(Windows/Linux) - ⏭️ Or use the play button in the toolbar above
- ⬇️ Move to the next cell and repeat
🧩 What happens when you run a cell?
The computer will either:
- ✏️ Show you some text or figures
- 🖥️ Run an analysis step in the background
- 📂 Save results into a folder
💡 Tip: Always read the explanation
There are a few dataformats that are used in this tutorial:
Files that end in ‘.mgz’ and ‘nii.gz’ are volumetric images (either showing the brain or a specific region of the brain).
Files that end in ‘.stat’ are statistics files.
Files that end in .txt are just text-files.
Files that end in ‘.csv’ are datasheets.
Files that end in ‘.pial, .surf etc’ are surface files.
Load the module for this workbook#
📦 What is a "Module"?
A module is just a way of telling the computer: “Please make this special software ready to use.”
- 🎛️ We only load the ones we need, when we need them
- 🔑 Loading a module is like unlocking the tool so we can use it
🚀 In this notebook
We will load FreeSurfer 8 – the software that analyses our scans. Once loaded, the commands will be available for us to run.
module load freesurfer/8.0.0
➡️ Just like before: click the cell, then press Command + Enter (Mac) or Control + Enter (Windows/Linux) to load it.
# we can use module to load freesurfer in a specific version
import module
await module.load('freesurfer/8.0.0')
await module.list()
['freesurfer/8.0.0']
%%capture
!pip install nibabel numpy
Let’s try running the command with the –help flag!#
!recon-all-clinical.sh --help
Recon-all-like stream for clinical scans of arbigrary orientation/resolution/contrast
Use this script to process clinical scans of arbitrary orientation, resolution, and
contrast. It essentially runs a combination of:
* SynthSeg: to obtain a volumetric segmentation and linear registration to Talairach space
* SynthSR: to have a higher resolution 1mm MPRAGE for visualization
* SynthDist: to fit surfaces by predicting the distance maps and reconstructing topologically accurate cortical surfaces
Using this module is very simple: you just provide an input scan, the subject name, the
number of threads you want to use, and (optionally) the subjects directory:
recon-all-clinical.sh INPUT_SCAN SUBJECT_ID THREADS [SUBJECT_DIR]
(the argument [SUBJECT_DIR] is only necessary if the
environment variable SUBJECTS_DIR has not been set
or if you want to override it)
This stream runs a bit faster than the original recon-all, since the volumetric
segmentation is much faster than the iterative Bayesian method in the standard stream
If you use this stream for your analysis, please cite:
K Gopinath, DN Greeve, S Das, S Arnold, C Magdamo, JE Iglesias:
Cortical analysis of heterogeneous clinical brain MRI scans for large-scale neuroimaging studies
https://arxiv.org/abs/2305.01827
B Billot, DN Greve, O Puonti, A Thielscher, K Van Leemput, B Fischl, AV Dalca, JE Iglesias:
SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining
Medical Image Analysis, 83, 102789 (2023)
B Billot, C Magdamo, SE Arnold, S Das, JE Iglesias:
Robust machine learning segmentation for large-scale analysis of heterogeneous clinical brain MRI datasets
PNAS, 120(9), e2216399120 (2023)
SynthSR: a public AI tool to turn heterogeneous clinical brain scans into high-resolution T1-weighted images for 3D morphometry
JE Iglesias, B Billot, Y Balbastre, C Magdamo, S Arnold, S Das, B Edlow, D Alexander, P Golland, B Fischl
Science Advances, 9(5), eadd3607 (2023)
📥 Downloading the data
We will now download a sample brain scan (called mp2rage.nii.gz)
from the Open Science Framework.
- 🌐 This file comes from an open research resource
- 🧠 It is a standard type of MRI scan used for brain structure
- 📂 We will use it today as our example dataset
➡️ Click the cell below, then press Command + Enter (Mac) or Control + Enter (Windows/Linux)
to start the download.
! echo "Downloading File"; mkdir -p ./data/structural && osf -p bt4ez fetch osfstorage/TOMCAT_DIB/sub-01/ses-01_7T/anat/sub-01_ses-01_7T_T1w_defaced.nii.gz ./data/structural/mp2rage.nii.gz
Downloading File
100%|██████████████████████████████████| 72.7M/72.7M [00:08<00:00, 9.06Mbytes/s]
!ls ./data/structural/mp*
./data/structural/mp2rage.nii.gz
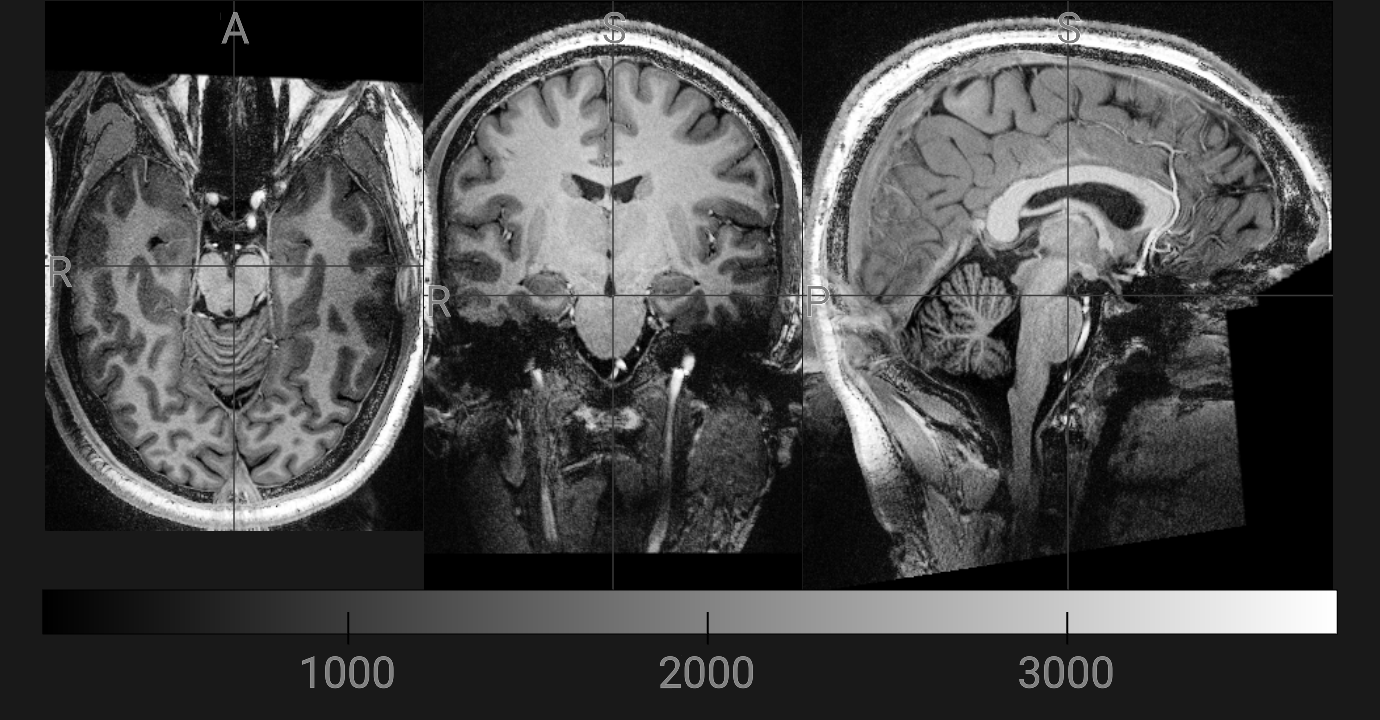
🖼️ Viewing mp2rage.nii.gz
We will now open the mp2rage.nii.gz file. This is the patient’s T1-weighted MRI scan that FreeSurfer will change into its own format.
- 🎛️ We display it in grayscale so brain tissue boundaries are clear
- 📐 Brightness/contrast is automatically adjusted (ignores empty space and extreme values)
- ➕ Crosshairs help you line up the same point across axial, coronal, and sagittal slices
- 🧭 The viewer lets you scroll through slices and explore the brain interactively
➡️ Click the cell below, then press Command + Enter (Mac) or Control + Enter (Windows/Linux).
This will open mp2rage.nii.gz in the interactive viewer.
# Display original file in grayscale
from ipyniivue import NiiVue
from IPython.display import display, Image
import nibabel as nib, numpy as np
T1_PATH = "./data/structural/mp2rage.nii.gz"
# Robust intensity window (ignore zeros and outliers)
t1 = nib.load(T1_PATH).get_fdata()
vals = t1[t1 > 0]
vmin, vmax = np.percentile(vals, [0.5, 99.5]) if vals.size else (None, None)
nv = NiiVue(
height=600,
multiplanar_layout="GRID",
multiplanar_show_render=True,
is_ruler=False,
is_colorbar=True,
is_orient_cube=True,
is_radiological_convention=True,
back_color=(0.1,0.1,0.1,1.0),
)
nv.add_volume({
"path": T1_PATH,
"name": "native",
"opacity": 1.0,
"colormap": "gray",
"cal_min": float(vmin) if vmin is not None else None,
"cal_max": float(vmax) if vmax is not None else None,
"ignore_zero_voxels": True,
})
nv.set_crosshair_color([0,0,0,1])
display(nv)
Image(url='https://raw.githubusercontent.com/neurodesk/example-notebooks/refs/heads/main/books/images/recon_all_clinical_t1w.png')
Run#
🧠 Automated Brain Scan Processing using recon-all clinical
This step prepares our MRI data so it can be processed automatically. Think of it like setting up the folders and tools before we start the real analysis.
- ✔️ Creates a folder where results will be saved
- ✔️ Checks everything is ready before running
- ✔️ Run the command "recon-all-clinical.sh"
In plain words: we are telling the computer:
"Here’s the scan. Put the results in this folder. Run the pipeline."
⚠️ Don’t worry about the code details – they’re just making sure the computer behaves!
➡️ When you are ready, hit Command + Enter (Mac) or Control + Enter (Windows/Linux),
or press the ▶️ play button after clicking on the cell below.
%%bash
#--------------------------------------------
# FreeSurfer recon-all-clinical pipeline
#--------------------------------------------
set -euo pipefail # Stop on unset variables or errors
#----------------------------
# Config
#----------------------------
INPUT_SCAN=./data/structural/mp2rage.nii.gz
SUBJECT_ID=TestSubject
THREADS=2 #adapt if more resources are available
SUBJECT_DIR=./data/structural/recon-all-clinical
# Ensure output folder exists
mkdir -p "$SUBJECT_DIR"
echo "SUBJECT_DIR=$SUBJECT_DIR"
echo "SUBJECT_ID=$SUBJECT_ID"
echo "INPUT_SCAN=$INPUT_SCAN"
echo
#----------------------------
# Allow deeper folder structures in some FreeSurfer builds
#----------------------------
export FS_ALLOW_DEEP=1
export SINGULARITYENV_FS_ALLOW_DEEP=1
export APPTAINERENV_FS_ALLOW_DEEP=1
#----------------------------
# Start pipeline
#----------------------------
# Remove previous "is-running" file if it exists
rm -f "$SUBJECT_DIR/$SUBJECT_ID/scripts/IsRunning.lh+rh"
# Run the actual recon-all-clinical pipeline
recon-all-clinical.sh "$INPUT_SCAN" "$SUBJECT_ID" "$THREADS" "$SUBJECT_DIR"
#----------------------------
# End message
#----------------------------
echo "----- recon-all-clinical.sh completed -----"
echo "Results saved to: $SUBJECT_DIR/$SUBJECT_ID"
SUBJECT_DIR=./data/structural/recon-all-clinical
SUBJECT_ID=TestSubject
INPUT_SCAN=./data/structural/mp2rage.nii.gz
#--------------------------------------------
#@# recon-all-clinical Thu Oct 30 10:07:59 PM UTC 2025
mri_convert ./data/structural/mp2rage.nii.gz /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri/native.mgz
INFO: using NIfTI-1 sform (sform_code=1)
reading from ./data/structural/mp2rage.nii.gz...
TR=4310.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (0.999815, -0.0191912, -0.00103917)
j_ras = (0.019201, 0.999761, 0.0104718)
k_ras = (0.000837961, -0.0104898, 0.999945)
writing to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri/native.mgz...
SynthSeg-robust 2.0
using CPU, hiding all CUDA_VISIBLE_DEVICES
using 2 threads
addctab True
predicting 1/1
1/1 [==============================] - 284s 284s/step
segmentation saved in: /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri/synthseg.mgz
volumes saved in: /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/stats/synthseg.vol.csv
QC scores saved in: /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/stats/synthseg.qc.csv
#@# mri_synthseg vmpeak 53443556
If you use this tool in a publication, please cite:
SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining
B. Billot, D.N. Greve, O. Puonti, A. Thielscher, K. Van Leemput, B. Fischl, A.V. Dalca, J.E. Iglesias
Medical Image Analysis, 2023.
Robust machine learning segmentation for large-scale analysis of heterogeneous clinical brain MRI datasets
B. Billot, C. Magdamo, Y. Cheng, S.E. Arnold, S. Das, J.E. Iglesias
PNAS, 2023.
using CPU, hiding all CUDA_VISIBLE_DEVICES
Using general model from January 2023 (version 2)
/opt/freesurfer-8.0.0/models/synthsr_v20_230130.h5
using 2 threads
predicting 1/1
Prediction without flipping
1/1 [==============================] - 44s 44s/step
Prediction with flipping
1/1 [==============================] - 41s 41s/step
prediction saved in: /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri/synthSR.raw.mgz
If you use this tool in a publication, please cite:
Joint super-resolution and synthesis of 1 mm isotropic MP-RAGE volumes from clinical
MRI exams with scans of different orientation, resolution and contrast
JE Iglesias, B Billot, Y Balbastre, A Tabari, J Conklin, RG Gonzalez, DC Alexander,
P Golland, BL Edlow, B Fischl, for the ADNI
NeuroImage, 118206 (2021)
SynthSR: a public AI tool to turn heterogeneous clinical brain scans into
high-resolution T1-weighted images for 3D morphometry
JE Iglesias, B Billot, Y Balbastre, C Magdamo, S Arnold, S Das, B Edlow, D Alexander,
P Golland, B Fischl
Science Advances, 9(5), eadd3607 (2023)
If you use the low-field (Hyperfine) version, please cite also:
Quantitative Brain Morphometry of Portable Low-Field-Strength MRI Using
Super-Resolution Machine Learning
JE Iglesias, R Schleicher, S Laguna, B Billot, P Schaefer, B McKaig, JN Goldstein,
KN Sheth, MS Rosen, WT Kimberly
Radiology, 220522 (2022)
2025-10-30 22:15:26.352249: I tensorflow/tsl/cuda/cudart_stub.cc:28] Could not find cuda drivers on your machine, GPU will not be used.
2025-10-30 22:15:26.422030: I tensorflow/tsl/cuda/cudart_stub.cc:28] Could not find cuda drivers on your machine, GPU will not be used.
2025-10-30 22:15:26.422803: I tensorflow/core/platform/cpu_feature_guard.cc:182] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.
To enable the following instructions: AVX2 AVX512F FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.
2025-10-30 22:15:27.619467: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT
/opt/freesurfer-8.0.0/python
using CPU, hiding all CUDA_VISIBLE_DEVICES
Reading image
Reading SynthSeg segmentation / parcellation
Resampling and padding image
Building U-net
Pushing image through U-net
1/1 [==============================] - 42s 42s/step
/opt/freesurfer-8.0.0/python/scripts/mri_synth_surf.py:12: DeprecationWarning: Please use `distance_transform_edt` from the `scipy.ndimage` namespace, the `scipy.ndimage.morphology` namespace is deprecated.
from scipy.ndimage.morphology import distance_transform_edt
Saving prediction to disk
Resampling SynthSeg to space of prediction
Computing Talairach transform
Making synthetic image and other <<fake>> volumes, in FreeSurfer orientation!
All done!
mri_convert ./synthseg.mgz ./synthseg.resampled.mgz -rl ./synthSR.raw.mgz -rt nearest -odt float
reading from ./synthseg.mgz...
TR=0.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (0.999815, -0.0191912, -0.00103917)
j_ras = (0.019201, 0.999761, 0.0104718)
k_ras = (0.000837961, -0.0104898, 0.999945)
reading template info from volume ./synthSR.raw.mgz...
writing to ./synthseg.resampled.mgz...
/opt/freesurfer-8.0.0/python/scripts/norm_synthSR.py:8: DeprecationWarning: Please use `distance_transform_edt` from the `scipy.ndimage` namespace, the `scipy.ndimage.morphology` namespace is deprecated.
from scipy.ndimage.morphology import distance_transform_edt
/opt/freesurfer-8.0.0/python
Reading inputs
Normalizing and dilating
Writing to disk
Done!
mri_convert ./synthSR.norm.tmp.mgz ./synthSR.norm.mgz -rl ./norm.mgz -odt float
reading from ./synthSR.norm.tmp.mgz...
TR=0.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (0.999815, -0.0191912, -0.00103917)
j_ras = (0.019201, 0.999761, 0.0104718)
k_ras = (0.000837961, -0.0104898, 0.999945)
reading template info from volume ./norm.mgz...
Reslicing using trilinear interpolation
writing to ./synthSR.norm.mgz...
8.0.0-beta
--src: norm.mgz src image (geometry).
--trg: /opt/freesurfer-8.0.0/average/mni305.cor.mgz trg image (geometry).
--inmni: transforms/talairach.xfm input MNI/XFM transform.
--outlta: transforms/talairach.xfm.lta output LTA.
--s: fsaverage subject name
--ltavox2vox: output LTA as VOX_TO_VOX transform.
LTA read, type : 1
1.10460 -0.00862 -0.00334 -0.03559;
-0.00180 1.03584 0.25272 -35.48325;
-0.01409 -0.29788 1.10985 47.01420;
0.00000 0.00000 0.00000 1.00000;
setting subject to fsaverage
Writing LTA to file transforms/talairach.xfm.lta...
lta_convert successful.
will read norm from ./synthSR.norm.mgz
will read input aseg from ./aseg.auto_noCCseg.mgz
writing aseg with cc labels to aseg.presurf.mgz
will write lta as ./transforms/cc_up.lta
reading aseg from /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri/./aseg.auto_noCCseg.mgz
reading norm from /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri/./synthSR.norm.mgz
195958 voxels in left wm, 50361 in right wm, xrange [72, 97]
searching rotation angles z=[-5 9], y=[-8 6]
searching scale 1 Z rot 8.5 global minimum found at slice 76.0, rotations (-0.83, 1.72)
final transformation (x=76.0, yr=-0.827, zr=1.720):
0.99945 -0.03002 -0.01442 55.66253;
0.03001 0.99955 -0.00043 51.79436;
0.01443 -0.00000 0.99990 29.91334;
0.00000 0.00000 0.00000 1.00000;
updating x range to be [125, 130] in xformed coordinates
best xformed slice 127
min_x_fornix = 124
min_x_fornix = 127
min_x_fornix = 134
min_x_fornix = 154
min_x_fornix = 123
cc center is found at 127 74 97
eigenvectors:
0.00040 -0.00484 0.99999;
-0.08493 -0.99638 -0.00478;
0.99639 -0.08492 -0.00081;
error in mid anterior detected - correcting...
writing aseg with callosum to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri/aseg.presurf.mgz...
corpus callosum segmentation took 1.1 minutes
#VMPC# mri_cc VmPeak 246464
mri_cc done
Iteration Number : 1
pass 1 (xy+): 37 found - 37 modified | TOTAL: 37
pass 2 (xy+): 0 found - 37 modified | TOTAL: 37
pass 1 (xy-): 34 found - 34 modified | TOTAL: 71
pass 2 (xy-): 0 found - 34 modified | TOTAL: 71
pass 1 (yz+): 54 found - 54 modified | TOTAL: 125
pass 2 (yz+): 0 found - 54 modified | TOTAL: 125
pass 1 (yz-): 52 found - 52 modified | TOTAL: 177
pass 2 (yz-): 0 found - 52 modified | TOTAL: 177
pass 1 (xz+): 36 found - 36 modified | TOTAL: 213
pass 2 (xz+): 0 found - 36 modified | TOTAL: 213
pass 1 (xz-): 41 found - 41 modified | TOTAL: 254
pass 2 (xz-): 0 found - 41 modified | TOTAL: 254
Iteration Number : 1
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 2 found - 2 modified | TOTAL: 2
pass 2 (+++): 0 found - 2 modified | TOTAL: 2
pass 1 (+++): 0 found - 0 modified | TOTAL: 2
pass 1 (+++): 4 found - 4 modified | TOTAL: 6
pass 2 (+++): 0 found - 4 modified | TOTAL: 6
Iteration Number : 1
pass 1 (++): 23 found - 23 modified | TOTAL: 23
pass 2 (++): 0 found - 23 modified | TOTAL: 23
pass 1 (+-): 11 found - 11 modified | TOTAL: 34
pass 2 (+-): 0 found - 11 modified | TOTAL: 34
pass 1 (--): 15 found - 15 modified | TOTAL: 49
pass 2 (--): 0 found - 15 modified | TOTAL: 49
pass 1 (-+): 27 found - 27 modified | TOTAL: 76
pass 2 (-+): 0 found - 27 modified | TOTAL: 76
Iteration Number : 2
pass 1 (xy+): 2 found - 2 modified | TOTAL: 2
pass 2 (xy+): 0 found - 2 modified | TOTAL: 2
pass 1 (xy-): 1 found - 1 modified | TOTAL: 3
pass 2 (xy-): 0 found - 1 modified | TOTAL: 3
pass 1 (yz+): 0 found - 0 modified | TOTAL: 3
pass 1 (yz-): 0 found - 0 modified | TOTAL: 3
pass 1 (xz+): 0 found - 0 modified | TOTAL: 3
pass 1 (xz-): 0 found - 0 modified | TOTAL: 3
Iteration Number : 2
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 2
pass 1 (++): 1 found - 1 modified | TOTAL: 1
pass 2 (++): 0 found - 1 modified | TOTAL: 1
pass 1 (+-): 0 found - 0 modified | TOTAL: 1
pass 1 (--): 0 found - 0 modified | TOTAL: 1
pass 1 (-+): 0 found - 0 modified | TOTAL: 1
Iteration Number : 3
pass 1 (xy+): 0 found - 0 modified | TOTAL: 0
pass 1 (xy-): 0 found - 0 modified | TOTAL: 0
pass 1 (yz+): 0 found - 0 modified | TOTAL: 0
pass 1 (yz-): 0 found - 0 modified | TOTAL: 0
pass 1 (xz+): 0 found - 0 modified | TOTAL: 0
pass 1 (xz-): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Total Number of Modified Voxels = 340 (out of 553619: 0.061414)
binarizing input wm segmentation...
Ambiguous edge configurations...
mri_pretess done
Iteration Number : 1
pass 1 (xy+): 15 found - 15 modified | TOTAL: 15
pass 2 (xy+): 0 found - 15 modified | TOTAL: 15
pass 1 (xy-): 17 found - 17 modified | TOTAL: 32
pass 2 (xy-): 0 found - 17 modified | TOTAL: 32
pass 1 (yz+): 29 found - 29 modified | TOTAL: 61
pass 2 (yz+): 0 found - 29 modified | TOTAL: 61
pass 1 (yz-): 28 found - 28 modified | TOTAL: 89
pass 2 (yz-): 0 found - 28 modified | TOTAL: 89
pass 1 (xz+): 21 found - 21 modified | TOTAL: 110
pass 2 (xz+): 0 found - 21 modified | TOTAL: 110
pass 1 (xz-): 18 found - 18 modified | TOTAL: 128
pass 2 (xz-): 0 found - 18 modified | TOTAL: 128
Iteration Number : 1
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 2 found - 2 modified | TOTAL: 2
pass 2 (+++): 0 found - 2 modified | TOTAL: 2
pass 1 (+++): 0 found - 0 modified | TOTAL: 2
pass 1 (+++): 4 found - 4 modified | TOTAL: 6
pass 2 (+++): 0 found - 4 modified | TOTAL: 6
Iteration Number : 1
pass 1 (++): 7 found - 7 modified | TOTAL: 7
pass 2 (++): 0 found - 7 modified | TOTAL: 7
pass 1 (+-): 8 found - 8 modified | TOTAL: 15
pass 2 (+-): 0 found - 8 modified | TOTAL: 15
pass 1 (--): 6 found - 6 modified | TOTAL: 21
pass 2 (--): 0 found - 6 modified | TOTAL: 21
pass 1 (-+): 12 found - 12 modified | TOTAL: 33
pass 2 (-+): 0 found - 12 modified | TOTAL: 33
Iteration Number : 2
pass 1 (xy+): 1 found - 1 modified | TOTAL: 1
pass 2 (xy+): 0 found - 1 modified | TOTAL: 1
pass 1 (xy-): 1 found - 1 modified | TOTAL: 2
pass 2 (xy-): 0 found - 1 modified | TOTAL: 2
pass 1 (yz+): 0 found - 0 modified | TOTAL: 2
pass 1 (yz-): 0 found - 0 modified | TOTAL: 2
pass 1 (xz+): 0 found - 0 modified | TOTAL: 2
pass 1 (xz-): 0 found - 0 modified | TOTAL: 2
Iteration Number : 2
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 2
pass 1 (++): 1 found - 1 modified | TOTAL: 1
pass 2 (++): 0 found - 1 modified | TOTAL: 1
pass 1 (+-): 0 found - 0 modified | TOTAL: 1
pass 1 (--): 0 found - 0 modified | TOTAL: 1
pass 1 (-+): 0 found - 0 modified | TOTAL: 1
Iteration Number : 3
pass 1 (xy+): 0 found - 0 modified | TOTAL: 0
pass 1 (xy-): 0 found - 0 modified | TOTAL: 0
pass 1 (yz+): 0 found - 0 modified | TOTAL: 0
pass 1 (yz-): 0 found - 0 modified | TOTAL: 0
pass 1 (xz+): 0 found - 0 modified | TOTAL: 0
pass 1 (xz-): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Total Number of Modified Voxels = 170 (out of 279688: 0.060782)
Ambiguous edge configurations...
mri_pretess done
8.0.0-beta
8.0.0-beta
slice 30: 1793 vertices, 1909 faces
slice 40: 6521 vertices, 6718 faces
slice 50: 13667 vertices, 13967 faces
slice 60: 22813 vertices, 23150 faces
slice 70: 33144 vertices, 33506 faces
slice 80: 43432 vertices, 43777 faces
slice 90: 52552 vertices, 52892 faces
slice 100: 61974 vertices, 62320 faces
slice 110: 71377 vertices, 71717 faces
slice 120: 80466 vertices, 80812 faces
slice 130: 89203 vertices, 89508 faces
slice 140: 96262 vertices, 96514 faces
slice 150: 102723 vertices, 102927 faces
slice 160: 107904 vertices, 108093 faces
slice 170: 111920 vertices, 112062 faces
slice 180: 114420 vertices, 114473 faces
slice 190: 114564 vertices, 114582 faces
slice 200: 114564 vertices, 114582 faces
slice 210: 114564 vertices, 114582 faces
using the conformed surface RAS to save vertex points...
writing ../surf/lh.orig.nofix
using vox2ras matrix:
-1.00000 0.00000 0.00000 77.50000;
0.00000 0.00000 1.00000 -106.00000;
0.00000 -1.00000 0.00000 117.50000;
0.00000 0.00000 0.00000 1.00000;
counting number of connected components...
114564 voxel in cpt #1: X=-18 [v=114564,e=343746,f=229164] located at (-26.720881, -10.154708, 34.414169)
For the whole surface: X=-18 [v=114564,e=343746,f=229164]
One single component has been found
nothing to do
done
Iteration Number : 1
pass 1 (xy+): 23 found - 23 modified | TOTAL: 23
pass 2 (xy+): 0 found - 23 modified | TOTAL: 23
pass 1 (xy-): 18 found - 18 modified | TOTAL: 41
pass 2 (xy-): 0 found - 18 modified | TOTAL: 41
pass 1 (yz+): 35 found - 35 modified | TOTAL: 76
pass 2 (yz+): 0 found - 35 modified | TOTAL: 76
pass 1 (yz-): 28 found - 28 modified | TOTAL: 104
pass 2 (yz-): 0 found - 28 modified | TOTAL: 104
pass 1 (xz+): 15 found - 15 modified | TOTAL: 119
pass 2 (xz+): 0 found - 15 modified | TOTAL: 119
pass 1 (xz-): 23 found - 23 modified | TOTAL: 142
pass 2 (xz-): 0 found - 23 modified | TOTAL: 142
Iteration Number : 1
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 1
pass 1 (++): 16 found - 16 modified | TOTAL: 16
pass 2 (++): 0 found - 16 modified | TOTAL: 16
pass 1 (+-): 4 found - 4 modified | TOTAL: 20
pass 2 (+-): 0 found - 4 modified | TOTAL: 20
pass 1 (--): 9 found - 9 modified | TOTAL: 29
pass 2 (--): 0 found - 9 modified | TOTAL: 29
pass 1 (-+): 14 found - 14 modified | TOTAL: 43
pass 2 (-+): 0 found - 14 modified | TOTAL: 43
Iteration Number : 2
pass 1 (xy+): 1 found - 1 modified | TOTAL: 1
pass 2 (xy+): 0 found - 1 modified | TOTAL: 1
pass 1 (xy-): 0 found - 0 modified | TOTAL: 1
pass 1 (yz+): 0 found - 0 modified | TOTAL: 1
pass 1 (yz-): 0 found - 0 modified | TOTAL: 1
pass 1 (xz+): 0 found - 0 modified | TOTAL: 1
pass 1 (xz-): 0 found - 0 modified | TOTAL: 1
Iteration Number : 2
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 2
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (xy+): 0 found - 0 modified | TOTAL: 0
pass 1 (xy-): 0 found - 0 modified | TOTAL: 0
pass 1 (yz+): 0 found - 0 modified | TOTAL: 0
pass 1 (yz-): 0 found - 0 modified | TOTAL: 0
pass 1 (xz+): 0 found - 0 modified | TOTAL: 0
pass 1 (xz-): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Total Number of Modified Voxels = 186 (out of 273947: 0.067896)
Ambiguous edge configurations...
mri_pretess done
8.0.0-beta
8.0.0-beta
slice 30: 2064 vertices, 2185 faces
slice 40: 7248 vertices, 7486 faces
slice 50: 14919 vertices, 15232 faces
slice 60: 24127 vertices, 24453 faces
slice 70: 33564 vertices, 33902 faces
slice 80: 42847 vertices, 43201 faces
slice 90: 51993 vertices, 52313 faces
slice 100: 60965 vertices, 61292 faces
slice 110: 69472 vertices, 69808 faces
slice 120: 78189 vertices, 78543 faces
slice 130: 86910 vertices, 87215 faces
slice 140: 94287 vertices, 94522 faces
slice 150: 100523 vertices, 100751 faces
slice 160: 106084 vertices, 106308 faces
slice 170: 110745 vertices, 110908 faces
slice 180: 113440 vertices, 113518 faces
slice 190: 113640 vertices, 113664 faces
slice 200: 113640 vertices, 113664 faces
slice 210: 113640 vertices, 113664 faces
using the conformed surface RAS to save vertex points...
writing ../surf/rh.orig.nofix
using vox2ras matrix:
-1.00000 0.00000 0.00000 77.50000;
0.00000 0.00000 1.00000 -106.00000;
0.00000 -1.00000 0.00000 117.50000;
0.00000 0.00000 0.00000 1.00000;
counting number of connected components...
113632 voxel in cpt #1: X=-26 [v=113632,e=340974,f=227316] located at (28.477621, -9.755870, 33.786083)
8 voxel in cpt #2: X=2 [v=8,e=18,f=12] located at (18.500000, -61.000000, 10.500000)
For the whole surface: X=-24 [v=113640,e=340992,f=227328]
2 components have been found
keeping component #1 with 113632 vertices
done
setting seed for random number generator to 1234
smoothing surface tessellation for 10 iterations...
smoothing complete - recomputing first and second fundamental forms...
setting seed for random number generator to 1234
smoothing surface tessellation for 10 iterations...
smoothing complete - recomputing first and second fundamental forms...
Not saving sulc
Reading ./lh.smoothwm.nofix
avg radius = 45.9 mm, total surface area = 62396 mm^2
step 060: RMS=0.026 (target=0.015) writing inflated surface to ./lh.inflated.nofix
inflation took 0.4 minutes
inflation complete.
Not saving sulc
mris_inflate utimesec 19.714050
mris_inflate stimesec 1.412997
mris_inflate ru_maxrss 182884
mris_inflate ru_ixrss 0
mris_inflate ru_idrss 0
mris_inflate ru_isrss 0
mris_inflate ru_minflt 478759
mris_inflate ru_majflt 16
mris_inflate ru_nswap 0
mris_inflate ru_inblock 3712
mris_inflate ru_oublock 8064
mris_inflate ru_msgsnd 0
mris_inflate ru_msgrcv 0
mris_inflate ru_nsignals 0
mris_inflate ru_nvcsw 52
mris_inflate ru_nivcsw 382
Not saving sulc
Reading ./rh.smoothwm.nofix
avg radius = 46.7 mm, total surface area = 61806 mm^2
step 060: RMS=0.028 (target=0.015) writing inflated surface to ./rh.inflated.nofix
inflation took 0.3 minutes
inflation complete.
Not saving sulc
mris_inflate utimesec 19.416371
mris_inflate stimesec 0.788821
mris_inflate ru_maxrss 181160
mris_inflate ru_ixrss 0
mris_inflate ru_idrss 0
mris_inflate ru_isrss 0
mris_inflate ru_minflt 233136
mris_inflate ru_majflt 0
mris_inflate ru_nswap 0
mris_inflate ru_inblock 0
mris_inflate ru_oublock 8000
mris_inflate ru_msgsnd 0
mris_inflate ru_msgrcv 0
mris_inflate ru_nsignals 0
mris_inflate ru_nvcsw 28
mris_inflate ru_nivcsw 140
doing quick spherical unfolding.
limitting unfolding to 6 passes
using n_averages = 128
setting seed for random number genererator to 1234
version: 8.0.0-beta
available threads: 1
scaling brain by 0.339...
inflating...
projecting onto sphere...
surface projected - minimizing metric distortion...
vertex spacing 1.08 +- 0.57 (0.02-->5.32) (max @ vno 70048 --> 70070)
face area 0.03 +- 0.03 (-0.09-->0.42)
Entering MRISinflateToSphere()
inflating to sphere (rms error < 2.00)
000: dt: 0.0000, rms radial error=176.315, avgs=0
005/300: dt: 0.9000, rms radial error=176.054, avgs=0
010/300: dt: 0.9000, rms radial error=175.492, avgs=0
015/300: dt: 0.9000, rms radial error=174.753, avgs=0
020/300: dt: 0.9000, rms radial error=173.913, avgs=0
025/300: dt: 0.9000, rms radial error=173.014, avgs=0
030/300: dt: 0.9000, rms radial error=172.083, avgs=0
035/300: dt: 0.9000, rms radial error=171.137, avgs=0
040/300: dt: 0.9000, rms radial error=170.187, avgs=0
045/300: dt: 0.9000, rms radial error=169.240, avgs=0
050/300: dt: 0.9000, rms radial error=168.295, avgs=0
055/300: dt: 0.9000, rms radial error=167.353, avgs=0
060/300: dt: 0.9000, rms radial error=166.416, avgs=0
065/300: dt: 0.9000, rms radial error=165.482, avgs=0
070/300: dt: 0.9000, rms radial error=164.554, avgs=0
075/300: dt: 0.9000, rms radial error=163.632, avgs=0
080/300: dt: 0.9000, rms radial error=162.715, avgs=0
085/300: dt: 0.9000, rms radial error=161.803, avgs=0
090/300: dt: 0.9000, rms radial error=160.896, avgs=0
095/300: dt: 0.9000, rms radial error=159.993, avgs=0
100/300: dt: 0.9000, rms radial error=159.096, avgs=0
105/300: dt: 0.9000, rms radial error=158.203, avgs=0
110/300: dt: 0.9000, rms radial error=157.316, avgs=0
115/300: dt: 0.9000, rms radial error=156.433, avgs=0
120/300: dt: 0.9000, rms radial error=155.555, avgs=0
125/300: dt: 0.9000, rms radial error=154.682, avgs=0
130/300: dt: 0.9000, rms radial error=153.814, avgs=0
135/300: dt: 0.9000, rms radial error=152.950, avgs=0
140/300: dt: 0.9000, rms radial error=152.092, avgs=0
145/300: dt: 0.9000, rms radial error=151.238, avgs=0
150/300: dt: 0.9000, rms radial error=150.389, avgs=0
155/300: dt: 0.9000, rms radial error=149.544, avgs=0
160/300: dt: 0.9000, rms radial error=148.705, avgs=0
165/300: dt: 0.9000, rms radial error=147.870, avgs=0
170/300: dt: 0.9000, rms radial error=147.039, avgs=0
175/300: dt: 0.9000, rms radial error=146.213, avgs=0
180/300: dt: 0.9000, rms radial error=145.392, avgs=0
185/300: dt: 0.9000, rms radial error=144.575, avgs=0
190/300: dt: 0.9000, rms radial error=143.763, avgs=0
195/300: dt: 0.9000, rms radial error=142.956, avgs=0
200/300: dt: 0.9000, rms radial error=142.152, avgs=0
205/300: dt: 0.9000, rms radial error=141.354, avgs=0
210/300: dt: 0.9000, rms radial error=140.559, avgs=0
215/300: dt: 0.9000, rms radial error=139.769, avgs=0
220/300: dt: 0.9000, rms radial error=138.984, avgs=0
225/300: dt: 0.9000, rms radial error=138.203, avgs=0
230/300: dt: 0.9000, rms radial error=137.426, avgs=0
235/300: dt: 0.9000, rms radial error=136.653, avgs=0
240/300: dt: 0.9000, rms radial error=135.885, avgs=0
245/300: dt: 0.9000, rms radial error=135.121, avgs=0
250/300: dt: 0.9000, rms radial error=134.361, avgs=0
255/300: dt: 0.9000, rms radial error=133.606, avgs=0
260/300: dt: 0.9000, rms radial error=132.855, avgs=0
265/300: dt: 0.9000, rms radial error=132.108, avgs=0
270/300: dt: 0.9000, rms radial error=131.365, avgs=0
275/300: dt: 0.9000, rms radial error=130.626, avgs=0
280/300: dt: 0.9000, rms radial error=129.891, avgs=0
285/300: dt: 0.9000, rms radial error=129.161, avgs=0
290/300: dt: 0.9000, rms radial error=128.434, avgs=0
295/300: dt: 0.9000, rms radial error=127.712, avgs=0
300/300: dt: 0.9000, rms radial error=126.994, avgs=0
spherical inflation complete.
epoch 1 (K=10.0), pass 1, starting sse = 12886.21
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.01/13 = 0.00054
epoch 2 (K=40.0), pass 1, starting sse = 1900.01
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.01/13 = 0.00076
epoch 3 (K=160.0), pass 1, starting sse = 139.83
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.19/17 = 0.01147
epoch 4 (K=640.0), pass 1, starting sse = 4.19
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.23/24 = 0.00962
final distance error %100000.00
writing spherical brain to ./lh.qsphere.nofix
spherical transformation took 0.0413 hours
FSRUNTIME@ mris_sphere 0.0413 hours 1 threads
#VMPC# mris_sphere VmPeak 443500
mris_sphere done
doing quick spherical unfolding.
limitting unfolding to 6 passes
using n_averages = 128
setting seed for random number genererator to 1234
version: 8.0.0-beta
available threads: 1
scaling brain by 0.340...
inflating...
projecting onto sphere...
surface projected - minimizing metric distortion...
vertex spacing 1.08 +- 0.58 (0.01-->5.47) (max @ vno 48028 --> 48890)
face area 0.03 +- 0.04 (-0.06-->0.54)
Entering MRISinflateToSphere()
inflating to sphere (rms error < 2.00)
000: dt: 0.0000, rms radial error=176.052, avgs=0
005/300: dt: 0.9000, rms radial error=175.792, avgs=0
010/300: dt: 0.9000, rms radial error=175.232, avgs=0
015/300: dt: 0.9000, rms radial error=174.499, avgs=0
020/300: dt: 0.9000, rms radial error=173.665, avgs=0
025/300: dt: 0.9000, rms radial error=172.774, avgs=0
030/300: dt: 0.9000, rms radial error=171.852, avgs=0
035/300: dt: 0.9000, rms radial error=170.914, avgs=0
040/300: dt: 0.9000, rms radial error=169.969, avgs=0
045/300: dt: 0.9000, rms radial error=169.024, avgs=0
050/300: dt: 0.9000, rms radial error=168.080, avgs=0
055/300: dt: 0.9000, rms radial error=167.139, avgs=0
060/300: dt: 0.9000, rms radial error=166.202, avgs=0
065/300: dt: 0.9000, rms radial error=165.273, avgs=0
070/300: dt: 0.9000, rms radial error=164.347, avgs=0
075/300: dt: 0.9000, rms radial error=163.427, avgs=0
080/300: dt: 0.9000, rms radial error=162.511, avgs=0
085/300: dt: 0.9000, rms radial error=161.600, avgs=0
090/300: dt: 0.9000, rms radial error=160.694, avgs=0
095/300: dt: 0.9000, rms radial error=159.793, avgs=0
100/300: dt: 0.9000, rms radial error=158.897, avgs=0
105/300: dt: 0.9000, rms radial error=158.005, avgs=0
110/300: dt: 0.9000, rms radial error=157.119, avgs=0
115/300: dt: 0.9000, rms radial error=156.237, avgs=0
120/300: dt: 0.9000, rms radial error=155.360, avgs=0
125/300: dt: 0.9000, rms radial error=154.488, avgs=0
130/300: dt: 0.9000, rms radial error=153.620, avgs=0
135/300: dt: 0.9000, rms radial error=152.757, avgs=0
140/300: dt: 0.9000, rms radial error=151.899, avgs=0
145/300: dt: 0.9000, rms radial error=151.045, avgs=0
150/300: dt: 0.9000, rms radial error=150.196, avgs=0
155/300: dt: 0.9000, rms radial error=149.351, avgs=0
160/300: dt: 0.9000, rms radial error=148.512, avgs=0
165/300: dt: 0.9000, rms radial error=147.677, avgs=0
170/300: dt: 0.9000, rms radial error=146.846, avgs=0
175/300: dt: 0.9000, rms radial error=146.021, avgs=0
180/300: dt: 0.9000, rms radial error=145.200, avgs=0
185/300: dt: 0.9000, rms radial error=144.383, avgs=0
190/300: dt: 0.9000, rms radial error=143.571, avgs=0
195/300: dt: 0.9000, rms radial error=142.763, avgs=0
200/300: dt: 0.9000, rms radial error=141.960, avgs=0
205/300: dt: 0.9000, rms radial error=141.162, avgs=0
210/300: dt: 0.9000, rms radial error=140.367, avgs=0
215/300: dt: 0.9000, rms radial error=139.577, avgs=0
220/300: dt: 0.9000, rms radial error=138.792, avgs=0
225/300: dt: 0.9000, rms radial error=138.011, avgs=0
230/300: dt: 0.9000, rms radial error=137.234, avgs=0
235/300: dt: 0.9000, rms radial error=136.462, avgs=0
240/300: dt: 0.9000, rms radial error=135.694, avgs=0
245/300: dt: 0.9000, rms radial error=134.930, avgs=0
250/300: dt: 0.9000, rms radial error=134.170, avgs=0
255/300: dt: 0.9000, rms radial error=133.414, avgs=0
260/300: dt: 0.9000, rms radial error=132.663, avgs=0
265/300: dt: 0.9000, rms radial error=131.916, avgs=0
270/300: dt: 0.9000, rms radial error=131.173, avgs=0
275/300: dt: 0.9000, rms radial error=130.435, avgs=0
280/300: dt: 0.9000, rms radial error=129.700, avgs=0
285/300: dt: 0.9000, rms radial error=128.970, avgs=0
290/300: dt: 0.9000, rms radial error=128.243, avgs=0
295/300: dt: 0.9000, rms radial error=127.521, avgs=0
300/300: dt: 0.9000, rms radial error=126.803, avgs=0
spherical inflation complete.
epoch 1 (K=10.0), pass 1, starting sse = 12774.46
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.01/13 = 0.00051
epoch 2 (K=40.0), pass 1, starting sse = 1888.78
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.01/13 = 0.00111
epoch 3 (K=160.0), pass 1, starting sse = 144.08
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.19/18 = 0.01042
epoch 4 (K=640.0), pass 1, starting sse = 6.14
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.17/25 = 0.00679
final distance error %100000.00
writing spherical brain to ./rh.qsphere.nofix
spherical transformation took 0.0441 hours
FSRUNTIME@ mris_sphere 0.0441 hours 1 threads
#VMPC# mris_sphere VmPeak 441924
mris_sphere done
reading spherical homeomorphism from 'qsphere.nofix'
reading inflated coordinates from 'inflated.nofix'
reading original coordinates from 'orig.nofix'
using genetic algorithm with optimized parameters
setting seed for random number genererator to 1234
*************************************************************
Topology Correction Parameters
retessellation mode: genetic search
number of patches/generation : 10
number of generations : 10
surface mri loglikelihood coefficient : 1.0
volume mri loglikelihood coefficient : 10.0
normal dot loglikelihood coefficient : 1.0
quadratic curvature loglikelihood coefficient : 1.0
volume resolution : 2
eliminate vertices during search : 1
initial patch selection : 1
select all defect vertices : 0
ordering dependant retessellation: 0
use precomputed edge table : 0
smooth retessellated patch : 2
match retessellated patch : 1
verbose mode : 0
*************************************************************
INFO: assuming .mgz format
writing corrected surface to 'orig.premesh'
8.0.0-beta
8.0.0-beta
before topology correction, eno=-18 (nv=114564, nf=229164, ne=343746, g=10)
using quasi-homeomorphic spherical map to tessellate cortical surface...
Correction of the Topology
Finding true center and radius of Spherical Surface...done
Surface centered at (0,0,0) with radius 100.0 in 11 iterations
marking ambiguous vertices...
807 ambiguous faces found in tessellation
segmenting defects...
12 defects found, arbitrating ambiguous regions...
analyzing neighboring defects...
12 defects to be corrected
0 vertices coincident
reading input surface /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/surf/lh.qsphere.nofix...
reading brain volume from brain...
reading wm segmentation from wm...
Reading original properties of orig.nofix
Reading vertex positions of inflated.nofix
Computing Initial Surface Statistics
-face loglikelihood: -9.3094 (-4.6547)
-vertex loglikelihood: -6.6149 (-3.3074)
-normal dot loglikelihood: -3.7133 (-3.7133)
-quad curv loglikelihood: -6.2532 (-3.1266)
Total Loglikelihood : -25.8907
CORRECTING DEFECT 0 (vertices=5, convex hull=31, v0=862)
After retessellation of defect 0 (v0=862), euler #=-9 (113994,341550,227547) : difference with theory (-9) = 0
CORRECTING DEFECT 1 (vertices=79, convex hull=112, v0=2342)
After retessellation of defect 1 (v0=2342), euler #=-8 (114004,341625,227613) : difference with theory (-8) = 0
CORRECTING DEFECT 2 (vertices=56, convex hull=104, v0=2502)
After retessellation of defect 2 (v0=2502), euler #=-7 (114013,341697,227677) : difference with theory (-7) = 0
CORRECTING DEFECT 3 (vertices=5, convex hull=35, v0=3807)
After retessellation of defect 3 (v0=3807), euler #=-6 (114015,341713,227692) : difference with theory (-6) = 0
CORRECTING DEFECT 4 (vertices=25, convex hull=81, v0=4670)
After retessellation of defect 4 (v0=4670), euler #=-5 (114024,341770,227741) : difference with theory (-5) = 0
CORRECTING DEFECT 5 (vertices=73, convex hull=111, v0=8988)
After retessellation of defect 5 (v0=8988), euler #=-4 (114059,341927,227864) : difference with theory (-4) = 0
CORRECTING DEFECT 6 (vertices=64, convex hull=109, v0=10458)
After retessellation of defect 6 (v0=10458), euler #=-3 (114086,342054,227965) : difference with theory (-3) = 0
CORRECTING DEFECT 7 (vertices=86, convex hull=139, v0=28658)
After retessellation of defect 7 (v0=28658), euler #=-2 (114137,342268,228129) : difference with theory (-2) = 0
CORRECTING DEFECT 8 (vertices=54, convex hull=83, v0=39107)
After retessellation of defect 8 (v0=39107), euler #=-1 (114170,342398,228227) : difference with theory (-1) = 0
CORRECTING DEFECT 9 (vertices=28, convex hull=76, v0=48957)
After retessellation of defect 9 (v0=48957), euler #=0 (114180,342461,228281) : difference with theory (0) = 0
CORRECTING DEFECT 10 (vertices=56, convex hull=97, v0=68226)
After retessellation of defect 10 (v0=68226), euler #=1 (114197,342555,228359) : difference with theory (1) = 0
CORRECTING DEFECT 11 (vertices=41, convex hull=77, v0=104704)
After retessellation of defect 11 (v0=104704), euler #=2 (114214,342636,228424) : difference with theory (2) = 0
computing original vertex metric properties...
storing new metric properties...
computing tessellation statistics...
vertex spacing 0.89 +- 0.21 (0.11-->5.86) (max @ vno 9519 --> 13195)
face area -nan +- -nan (1000.00-->-1.00)
performing soap bubble on retessellated vertices for 0 iterations...
vertex spacing 0.89 +- 0.21 (0.11-->5.86) (max @ vno 9519 --> 13195)
face area -nan +- -nan (1000.00-->-1.00)
tessellation finished, orienting corrected surface...
58 mutations (35.4%), 106 crossovers (64.6%), 102 vertices were eliminated
building final representation...
350 vertices and 0 faces have been removed from triangulation
after topology correction, eno=2 (nv=114214, nf=228424, ne=342636, g=0)
writing corrected surface to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/surf/lh.orig.premesh...
0.000 % of the vertices (0 vertices) exhibit an orientation change
removing intersecting faces
000: 75 intersecting
terminating search with 0 intersecting
topology fixing took 1.7 minutes
FSRUNTIME@ mris_fix_topology lh 0.0278 hours 1 threads
#VMPC# mris_fix_topology VmPeak 737756
reading spherical homeomorphism from 'qsphere.nofix'
reading inflated coordinates from 'inflated.nofix'
reading original coordinates from 'orig.nofix'
using genetic algorithm with optimized parameters
setting seed for random number genererator to 1234
*************************************************************
Topology Correction Parameters
retessellation mode: genetic search
number of patches/generation : 10
number of generations : 10
surface mri loglikelihood coefficient : 1.0
volume mri loglikelihood coefficient : 10.0
normal dot loglikelihood coefficient : 1.0
quadratic curvature loglikelihood coefficient : 1.0
volume resolution : 2
eliminate vertices during search : 1
initial patch selection : 1
select all defect vertices : 0
ordering dependant retessellation: 0
use precomputed edge table : 0
smooth retessellated patch : 2
match retessellated patch : 1
verbose mode : 0
*************************************************************
INFO: assuming .mgz format
writing corrected surface to 'orig.premesh'
8.0.0-beta
8.0.0-beta
before topology correction, eno=-26 (nv=113632, nf=227316, ne=340974, g=14)
using quasi-homeomorphic spherical map to tessellate cortical surface...
Correction of the Topology
Finding true center and radius of Spherical Surface...done
Surface centered at (0,0,0) with radius 100.0 in 8 iterations
marking ambiguous vertices...
2126 ambiguous faces found in tessellation
segmenting defects...
12 defects found, arbitrating ambiguous regions...
analyzing neighboring defects...
12 defects to be corrected
0 vertices coincident
reading input surface /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/surf/rh.qsphere.nofix...
reading brain volume from brain...
reading wm segmentation from wm...
Reading original properties of orig.nofix
Reading vertex positions of inflated.nofix
Computing Initial Surface Statistics
-face loglikelihood: -9.3162 (-4.6581)
-vertex loglikelihood: -6.5417 (-3.2709)
-normal dot loglikelihood: -3.6684 (-3.6684)
-quad curv loglikelihood: -6.2088 (-3.1044)
Total Loglikelihood : -25.7350
CORRECTING DEFECT 0 (vertices=312, convex hull=229, v0=15)
After retessellation of defect 0 (v0=15), euler #=-9 (112287,336293,223997) : difference with theory (-9) = 0
CORRECTING DEFECT 1 (vertices=76, convex hull=125, v0=1426)
After retessellation of defect 1 (v0=1426), euler #=-8 (112330,336473,224135) : difference with theory (-8) = 0
CORRECTING DEFECT 2 (vertices=139, convex hull=109, v0=1515)
After retessellation of defect 2 (v0=1515), euler #=-7 (112370,336652,224275) : difference with theory (-7) = 0
CORRECTING DEFECT 3 (vertices=139, convex hull=174, v0=3361)
After retessellation of defect 3 (v0=3361), euler #=-6 (112393,336799,224400) : difference with theory (-6) = 0
CORRECTING DEFECT 4 (vertices=127, convex hull=112, v0=13030)
After retessellation of defect 4 (v0=13030), euler #=-5 (112417,336925,224503) : difference with theory (-5) = 0
CORRECTING DEFECT 5 (vertices=55, convex hull=90, v0=15297)
After retessellation of defect 5 (v0=15297), euler #=-4 (112453,337068,224611) : difference with theory (-4) = 0
CORRECTING DEFECT 6 (vertices=70, convex hull=107, v0=17663)
After retessellation of defect 6 (v0=17663), euler #=-3 (112494,337235,224738) : difference with theory (-3) = 0
CORRECTING DEFECT 7 (vertices=45, convex hull=70, v0=19066)
After retessellation of defect 7 (v0=19066), euler #=-2 (112502,337289,224785) : difference with theory (-2) = 0
CORRECTING DEFECT 8 (vertices=191, convex hull=194, v0=55731)
After retessellation of defect 8 (v0=55731), euler #=-1 (112594,337658,225063) : difference with theory (-1) = 0
CORRECTING DEFECT 9 (vertices=16, convex hull=36, v0=60760)
After retessellation of defect 9 (v0=60760), euler #=0 (112598,337682,225084) : difference with theory (0) = 0
CORRECTING DEFECT 10 (vertices=123, convex hull=131, v0=98009)
After retessellation of defect 10 (v0=98009), euler #=1 (112650,337896,225247) : difference with theory (1) = 0
CORRECTING DEFECT 11 (vertices=75, convex hull=112, v0=102371)
After retessellation of defect 11 (v0=102371), euler #=2 (112665,337989,225326) : difference with theory (2) = 0
computing original vertex metric properties...
storing new metric properties...
computing tessellation statistics...
vertex spacing 0.89 +- 0.23 (0.08-->10.71) (max @ vno 88 --> 743)
face area -nan +- -nan (1000.00-->-1.00)
performing soap bubble on retessellated vertices for 0 iterations...
vertex spacing 0.89 +- 0.23 (0.08-->10.71) (max @ vno 88 --> 743)
face area -nan +- -nan (1000.00-->-1.00)
tessellation finished, orienting corrected surface...
49 mutations (32.7%), 101 crossovers (67.3%), 219 vertices were eliminated
building final representation...
967 vertices and 0 faces have been removed from triangulation
after topology correction, eno=2 (nv=112665, nf=225326, ne=337989, g=0)
writing corrected surface to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/surf/rh.orig.premesh...
0.000 % of the vertices (0 vertices) exhibit an orientation change
removing intersecting faces
000: 189 intersecting
001: 11 intersecting
terminating search with 0 intersecting
topology fixing took 2.6 minutes
FSRUNTIME@ mris_fix_topology rh 0.0441 hours 1 threads
#VMPC# mris_fix_topology VmPeak 736852
euler # = v-e+f = 2g-2: 114214 - 342636 + 228424 = 2 --> 0 holes
F =2V-4: 228424 = 228428-4 (0)
2E=3F: 685272 = 685272 (0)
total defect index = 0
euler # = v-e+f = 2g-2: 112665 - 337989 + 225326 = 2 --> 0 holes
F =2V-4: 225326 = 225330-4 (0)
2E=3F: 675978 = 675978 (0)
total defect index = 0
iters = 3
standard remeshing without target
adjusted l: 0.7099
remeshing to edge length 0.7099 with 3 iterations
avg qual before : 0.892138 after: 0.970858
Removing intersections
Remeshed surface quality stats nv0 = 114214 nv = 119790 1.04882
Area 239576 0.30137 0.03372 0.075405 0.4659
Corner 718728 60.00000 8.85952 15.843892 147.9050
Edge 359364 0.84230 0.08263 0.466086 1.2901
Hinge 359364 9.31013 9.25985 0.000000 113.8558
mris_remesh done
iters = 3
standard remeshing without target
adjusted l: 0.71114
remeshing to edge length 0.71114 with 3 iterations
avg qual before : 0.892023 after: 0.971275
Removing intersections
Remeshed surface quality stats nv0 = 112665 nv = 117922 1.04666
Area 235840 0.30290 0.03358 0.072959 0.4545
Corner 707520 60.00000 8.77844 18.154006 142.5635
Edge 353760 0.84432 0.08229 0.475901 1.2435
Hinge 353760 9.36595 9.41020 0.000000 119.1113
mris_remesh done
intersection removal took 0.00 hours
Found 0 intersections
writing corrected surface to ./lh.orig
intersection removal took 0.00 hours
Found 0 intersections
writing corrected surface to ./rh.orig
8.0.0-beta
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical
mris_autodet_gwstats --o ../surf/autodet.gw.stats.lh.dat --i brain.mgz --wm wm.mgz --surf ../surf/lh.orig.premesh
border white: 208355 voxels (2.70%)
border gray 225270 voxels (2.92%)
Reading in intensity volume brain.mgz
Reading in wm volume wm.mgz
Reading in surf ../surf/lh.orig.premesh
Auto detecting stats
MRIclipBrightWM(): nthresh=0, wmmin=5, clip=110
Binarizing thresholding at 5
computing class statistics... low=30, hi=110.000000
CCS WM (106.0): 100.9 +- 10.6 [70.0 --> 109.0]
CCS GM (70.0) : 73.0 +- 10.5 [30.0 --> 109.0]
white_mean = 100.922 +/- 10.583, gray_mean = 72.975 +/- 10.5025
using class modes intead of means, discounting robust sigmas....
MRIScomputeClassModes(): min=0 max=109 nbins=110
intensity peaks found at WM=108+-0.9, GM=69+-2.6
white_mode = 108, gray_mode = 69
std_scale = 1
Applying sanity checks, max_scale_down = 0.2
setting MIN_GRAY_AT_WHITE_BORDER to 58.5 (was 70.000000)
setting MAX_BORDER_WHITE to 118.6 (was 105.000000)
setting MIN_BORDER_WHITE to 69.0 (was 85.000000)
setting MAX_CSF to 48.0 (was 40.000000)
setting MAX_GRAY to 97.4 (was 95.000000)
setting MAX_GRAY_AT_CSF_BORDER to 58.5 (was 75.000000)
setting MIN_GRAY_AT_CSF_BORDER to 37.5 (was 40.000000)
When placing the white surface
white_border_hi = 118.583;
white_border_low = 69;
white_outside_low = 58.4975;
white_inside_hi = 120;
white_outside_hi = 118.583;
When placing the pial surface
pial_border_hi = 58.4975;
pial_border_low = 37.4926;
pial_outside_low = 10;
pial_inside_hi = 97.417;
pial_outside_hi = 53.2463;
#VMPC# mris_autodet_gwstats VmPeak 226108
mris_autodet_gwstats done
8.0.0-beta
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical
mris_autodet_gwstats --o ../surf/autodet.gw.stats.rh.dat --i brain.mgz --wm wm.mgz --surf ../surf/rh.orig.premesh
border white: 208355 voxels (2.70%)
border gray 225270 voxels (2.92%)
Reading in intensity volume brain.mgz
Reading in wm volume wm.mgz
Reading in surf ../surf/rh.orig.premesh
Auto detecting stats
MRIclipBrightWM(): nthresh=0, wmmin=5, clip=110
Binarizing thresholding at 5
computing class statistics... low=30, hi=110.000000
CCS WM (106.0): 100.9 +- 10.6 [70.0 --> 109.0]
CCS GM (70.0) : 73.0 +- 10.5 [30.0 --> 109.0]
white_mean = 100.922 +/- 10.583, gray_mean = 72.975 +/- 10.5025
using class modes intead of means, discounting robust sigmas....
MRIScomputeClassModes(): min=0 max=109 nbins=110
intensity peaks found at WM=108+-0.9, GM=69+-2.6
white_mode = 108, gray_mode = 69
std_scale = 1
Applying sanity checks, max_scale_down = 0.2
setting MIN_GRAY_AT_WHITE_BORDER to 58.5 (was 70.000000)
setting MAX_BORDER_WHITE to 118.6 (was 105.000000)
setting MIN_BORDER_WHITE to 69.0 (was 85.000000)
setting MAX_CSF to 48.0 (was 40.000000)
setting MAX_GRAY to 97.4 (was 95.000000)
setting MAX_GRAY_AT_CSF_BORDER to 58.5 (was 75.000000)
setting MIN_GRAY_AT_CSF_BORDER to 37.5 (was 40.000000)
When placing the white surface
white_border_hi = 118.583;
white_border_low = 69;
white_outside_low = 58.4975;
white_inside_hi = 120;
white_outside_hi = 118.583;
When placing the pial surface
pial_border_hi = 58.4975;
pial_border_low = 37.4926;
pial_outside_low = 10;
pial_inside_hi = 97.417;
pial_outside_hi = 53.2463;
#VMPC# mris_autodet_gwstats VmPeak 224000
mris_autodet_gwstats done
8.0.0-beta
8.0.0-beta
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical
mris_place_surface --adgws-in ../surf/autodet.gw.stats.lh.dat --wm wm.mgz --threads 2 --invol brain.mgz --lh --i ../surf/lh.orig --o ../surf/lh.white.preaparc --white --seg aseg.presurf.mgz --nsmooth 5
Reading in input surface ../surf/lh.orig
Smoothing surface before ripping with 5 iterations
Area 239576 0.26853 0.06118 0.029876 0.5937
Corner 718728 60.00000 9.11475 19.463542 134.0574
Edge 359364 0.79110 0.11071 0.179458 1.3129
Hinge 359364 6.35377 6.02304 0.000018 99.5614
Not reading in aparc
Reading in input volume brain.mgz
Reading in seg volume aseg.presurf.mgz
Reading in wm volume wm.mgz
MRIclipBrightWM(): nthresh=0, wmmin=5, clip=110
MRIfindBrightNonWM(): 0 bright non-wm voxels segmented.
Masking bright non-wm for white surface
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=0
#FML# MRISripMidline(): nmarked=6244, nmarked2=0, nripped=6244
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247
MRISripSegs(): -2 2 0.5 ripped 0
opt weights int=0.2 loc=0 rep=5 nspr=0.3 curv=1
vertex 59895: xyz = (-49.7828,1.66613,32.6105) oxyz = (-49.7828,1.66613,32.6105) wxzy = (-49.7828,1.66613,32.6105) pxyz = (0,0,0)
CBVO Creating mask 119790
n_averages 4
Iteration 0 =========================================
n_averages=4, current_sigma=2
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6244
#FML# MRISripMidline(): nmarked=6244, nmarked2=0, nripped=6244
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 2;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=119790
Gdiag_no=-1
VMPeak = 891400
vno start=0, stop=119790
#SI# sigma=2 had to be increased for 391 vertices, nripped=6244
mean border=78.3, 565 (565) missing vertices, mean dist 0.3 [0.8 (%32.4)->0.8 (%67.6))]
%60 local maxima, %34 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.1013 min
Finding expansion regions
mean absolute distance = 0.83 +- 1.02
3675 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=2.0, host=jupyt, nav=4, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height 0.000000
momentum 0.000000
nbhd_size 0
max_nbrs 0
niterations 100
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 0
randomSeed 0
000: dt: 0.0000, sse=5838869.5, rms=15.895
001: dt: 0.5000, sse=3730758.0, rms=12.628 (20.554%)
002: dt: 0.5000, sse=2478716.2, rms=10.202 (19.209%)
003: dt: 0.5000, sse=1705069.6, rms=8.356 (18.099%)
004: dt: 0.5000, sse=1226227.2, rms=6.964 (16.662%)
005: dt: 0.5000, sse=935485.7, rms=5.958 (14.443%)
006: dt: 0.5000, sse=756701.2, rms=5.249 (11.893%)
007: dt: 0.5000, sse=649368.4, rms=4.764 (9.237%)
008: dt: 0.5000, sse=584869.2, rms=4.448 (6.636%)
009: dt: 0.5000, sse=553749.5, rms=4.247 (4.515%)
010: dt: 0.5000, sse=536312.0, rms=4.112 (3.181%)
011: dt: 0.5000, sse=507321.2, rms=4.016 (2.351%)
012: dt: 0.5000, sse=504614.6, rms=3.949 (1.665%)
013: dt: 0.5000, sse=486224.4, rms=3.895 (1.361%)
rms = 3.8569/3.8951, sse=491912.3/486224.4, time step reduction 1 of 3 to 0.250 0 1 1
014: dt: 0.5000, sse=491912.3, rms=3.857 (0.980%)
015: dt: 0.2500, sse=292942.7, rms=2.542 (34.097%)
016: dt: 0.2500, sse=256117.1, rms=2.168 (14.689%)
017: dt: 0.2500, sse=246758.5, rms=2.065 (4.748%)
018: dt: 0.2500, sse=236558.8, rms=1.989 (3.682%)
rms = 1.9558/1.9894, sse=235857.4/236558.7, time step reduction 2 of 3 to 0.125 0 0 1
019: dt: 0.2500, sse=235857.4, rms=1.956 (1.689%)
020: dt: 0.1250, sse=228605.2, rms=1.902 (2.741%)
rms = 1.8891/1.9022, sse=230899.6/228605.2, time step reduction 3 of 3 to 0.062 0 1 1
021: dt: 0.1250, sse=230899.6, rms=1.889 (0.690%)
maximum number of reductions reached, breaking from loop
positioning took 1.8 minutes
done positioning surface
Iteration 1 =========================================
n_averages=2, current_sigma=1
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6244
removing 3 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6433, nmarked2=0, nripped=6433
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 1;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=119790
Gdiag_no=-1
VMPeak = 1725612
vno start=0, stop=119790
#SI# sigma=1 had to be increased for 215 vertices, nripped=6433
mean border=84.9, 550 (459) missing vertices, mean dist -0.3 [0.4 (%90.0)->0.2 (%10.0))]
%81 local maxima, %13 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0667 min
Finding expansion regions
mean absolute distance = 0.35 +- 0.41
820 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=1.0, host=jupyt, nav=2, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=1704851.8, rms=8.178
022: dt: 0.5000, sse=803548.4, rms=5.184 (36.613%)
023: dt: 0.5000, sse=643726.4, rms=4.450 (14.148%)
rms = 4.4120/4.4503, sse=636812.9/643726.4, time step reduction 1 of 3 to 0.250 0 0 1
024: dt: 0.5000, sse=636812.9, rms=4.412 (0.861%)
025: dt: 0.2500, sse=345509.2, rms=2.583 (41.458%)
026: dt: 0.2500, sse=294540.0, rms=2.067 (19.977%)
027: dt: 0.2500, sse=278940.7, rms=1.945 (5.888%)
028: dt: 0.2500, sse=271174.3, rms=1.853 (4.741%)
rms = 1.8242/1.8530, sse=271880.5/271174.3, time step reduction 2 of 3 to 0.125 0 1 1
029: dt: 0.2500, sse=271880.5, rms=1.824 (1.554%)
rms = 1.7477/1.8242, sse=272929.6/271880.5, time step reduction 3 of 3 to 0.062 0 1 0
030: dt: 0.1250, sse=272929.6, rms=1.748 (4.193%)
maximum number of reductions reached, breaking from loop
positioning took 0.7 minutes
done positioning surface
Iteration 2 =========================================
n_averages=1, current_sigma=0.5
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6433
removing 3 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6473, nmarked2=0, nripped=6473
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 0.5;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=119790
Gdiag_no=-1
VMPeak = 1904324
vno start=0, stop=119790
#SI# sigma=0.5 had to be increased for 197 vertices, nripped=6473
mean border=87.9, 592 (441) missing vertices, mean dist -0.1 [0.1 (%84.0)->0.1 (%16.0))]
%90 local maxima, % 4 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0381 min
Finding expansion regions
mean absolute distance = 0.14 +- 0.22
661 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.5, host=jupyt, nav=1, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=545890.5, rms=3.921
031: dt: 0.5000, sse=533253.4, rms=3.799 (3.122%)
rms = 4.2111/3.7987, sse=589766.6/533253.4, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
032: dt: 0.2500, sse=335481.1, rms=2.498 (34.238%)
033: dt: 0.2500, sse=283216.4, rms=1.959 (21.575%)
034: dt: 0.2500, sse=277476.4, rms=1.816 (7.301%)
035: dt: 0.2500, sse=267582.2, rms=1.765 (2.836%)
rms = 1.7438/1.7646, sse=261234.5/267582.2, time step reduction 2 of 3 to 0.125 0 0 1
036: dt: 0.2500, sse=261234.5, rms=1.744 (1.175%)
037: dt: 0.1250, sse=254960.4, rms=1.674 (4.010%)
rms = 1.6715/1.6739, sse=255120.2/254960.4, time step reduction 3 of 3 to 0.062 0 1 1
038: dt: 0.1250, sse=255120.2, rms=1.671 (0.146%)
maximum number of reductions reached, breaking from loop
positioning took 0.7 minutes
done positioning surface
Iteration 3 =========================================
n_averages=0, current_sigma=0.25
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6473
removing 4 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6486, nmarked2=0, nripped=6486
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 0.25;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=119790
Gdiag_no=-1
VMPeak = 1983668
vno start=0, stop=119790
#SI# sigma=0.25 had to be increased for 175 vertices, nripped=6486
mean border=88.3, 655 (437) missing vertices, mean dist -0.0 [0.1 (%55.9)->0.1 (%44.1))]
%91 local maxima, % 3 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0248 min
Finding expansion regions
mean absolute distance = 0.09 +- 0.16
954 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.2, host=jupyt, nav=0, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=262385.8, rms=1.775
rms = 2.1586/1.7753, sse=302813.2/262385.8, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
039: dt: 0.2500, sse=248310.8, rms=1.576 (11.211%)
rms = 1.5456/1.5763, sse=247687.9/248310.8, time step reduction 2 of 3 to 0.125 0 0 1
040: dt: 0.2500, sse=247687.9, rms=1.546 (1.945%)
rms = 1.5324/1.5456, sse=261752.9/247687.9, time step reduction 3 of 3 to 0.062 0 1 1
041: dt: 0.1250, sse=261752.9, rms=1.532 (0.855%)
maximum number of reductions reached, breaking from loop
positioning took 0.3 minutes
done positioning surface
Removing intersections
#ET# mris_place_surface 3.89 minutes
Writing output to ../surf/lh.white.preaparc
#VMPC# mris_place_surfaces VmPeak 2051784
mris_place_surface done
8.0.0-beta
8.0.0-beta
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical
mris_place_surface --adgws-in ../surf/autodet.gw.stats.rh.dat --wm wm.mgz --threads 2 --invol brain.mgz --rh --i ../surf/rh.orig --o ../surf/rh.white.preaparc --white --seg aseg.presurf.mgz --nsmooth 5
Reading in input surface ../surf/rh.orig
Smoothing surface before ripping with 5 iterations
Area 235840 0.26931 0.06148 0.013425 0.5851
Corner 707520 60.00000 9.13941 20.029026 133.8117
Edge 353760 0.79227 0.11109 0.146651 1.2577
Hinge 353760 6.38664 6.11601 0.000011 116.3464
Not reading in aparc
Reading in input volume brain.mgz
Reading in seg volume aseg.presurf.mgz
Reading in wm volume wm.mgz
MRIclipBrightWM(): nthresh=0, wmmin=5, clip=110
MRIfindBrightNonWM(): 0 bright non-wm voxels segmented.
Masking bright non-wm for white surface
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6242, nmarked2=0, nripped=6242
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247
MRISripSegs(): -2 2 0.5 ripped 0
opt weights int=0.2 loc=0 rep=5 nspr=0.3 curv=1
vertex 58961: xyz = (34.8115,3.75511,51.0703) oxyz = (34.8115,3.75511,51.0703) wxzy = (34.8115,3.75511,51.0703) pxyz = (0,0,0)
CBVO Creating mask 117922
n_averages 4
Iteration 0 =========================================
n_averages=4, current_sigma=2
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6242
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6242, nmarked2=0, nripped=6242
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 2;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=117922
Gdiag_no=-1
VMPeak = 887188
vno start=0, stop=117922
#SI# sigma=2 had to be increased for 306 vertices, nripped=6242
mean border=78.3, 502 (502) missing vertices, mean dist 0.4 [0.7 (%33.3)->0.9 (%66.7))]
%63 local maxima, %31 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.1008 min
Finding expansion regions
mean absolute distance = 0.82 +- 0.96
3399 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=2.0, host=jupyt, nav=4, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height 0.000000
momentum 0.000000
nbhd_size 0
max_nbrs 0
niterations 100
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 1
randomSeed 0
000: dt: 0.0000, sse=5354776.0, rms=15.332
001: dt: 0.5000, sse=3289522.5, rms=11.928 (22.197%)
002: dt: 0.5000, sse=2077289.5, rms=9.370 (21.449%)
003: dt: 0.5000, sse=1382639.5, rms=7.516 (19.790%)
004: dt: 0.5000, sse=988276.1, rms=6.223 (17.204%)
005: dt: 0.5000, sse=765210.3, rms=5.353 (13.980%)
006: dt: 0.5000, sse=635152.6, rms=4.768 (10.925%)
007: dt: 0.5000, sse=563391.1, rms=4.392 (7.876%)
008: dt: 0.5000, sse=515540.0, rms=4.145 (5.642%)
009: dt: 0.5000, sse=492023.0, rms=4.009 (3.279%)
010: dt: 0.5000, sse=478022.6, rms=3.906 (2.553%)
rms = 3.8606/3.9063, sse=469828.4/478022.6, time step reduction 1 of 3 to 0.250 0 0 1
011: dt: 0.5000, sse=469828.4, rms=3.861 (1.170%)
012: dt: 0.2500, sse=287934.5, rms=2.564 (33.594%)
013: dt: 0.2500, sse=248640.5, rms=2.194 (14.425%)
014: dt: 0.2500, sse=238514.9, rms=2.081 (5.127%)
015: dt: 0.2500, sse=230140.0, rms=1.996 (4.124%)
rms = 1.9594/1.9956, sse=228880.3/230140.0, time step reduction 2 of 3 to 0.125 0 0 1
016: dt: 0.2500, sse=228880.3, rms=1.959 (1.810%)
017: dt: 0.1250, sse=227234.5, rms=1.904 (2.854%)
rms = 1.8880/1.9035, sse=221167.4/227234.5, time step reduction 3 of 3 to 0.062 0 0 1
018: dt: 0.1250, sse=221167.4, rms=1.888 (0.817%)
maximum number of reductions reached, breaking from loop
positioning took 1.5 minutes
done positioning surface
Iteration 1 =========================================
n_averages=2, current_sigma=1
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6242
removing 4 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6407, nmarked2=0, nripped=6407
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 1;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=117922
Gdiag_no=-1
VMPeak = 1704612
vno start=0, stop=117922
#SI# sigma=1 had to be increased for 193 vertices, nripped=6407
mean border=85.1, 506 (426) missing vertices, mean dist -0.3 [0.4 (%90.8)->0.2 (%9.2))]
%84 local maxima, %11 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0650 min
Finding expansion regions
mean absolute distance = 0.35 +- 0.38
1352 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=1.0, host=jupyt, nav=2, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=1730947.5, rms=8.339
019: dt: 0.5000, sse=807793.8, rms=5.262 (36.902%)
020: dt: 0.5000, sse=636081.7, rms=4.467 (15.109%)
021: dt: 0.5000, sse=612913.2, rms=4.374 (2.067%)
rms = 4.4378/4.3744, sse=626413.6/612913.2, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
022: dt: 0.2500, sse=369782.8, rms=2.871 (34.374%)
023: dt: 0.2500, sse=296359.0, rms=2.153 (25.008%)
024: dt: 0.2500, sse=277288.0, rms=1.927 (10.493%)
025: dt: 0.2500, sse=267265.1, rms=1.851 (3.959%)
rms = 1.8076/1.8506, sse=265935.3/267265.1, time step reduction 2 of 3 to 0.125 0 0 1
026: dt: 0.2500, sse=265935.3, rms=1.808 (2.327%)
027: dt: 0.1250, sse=259442.5, rms=1.728 (4.393%)
rms = 1.7183/1.7282, sse=263697.9/259442.5, time step reduction 3 of 3 to 0.062 0 1 1
028: dt: 0.1250, sse=263697.9, rms=1.718 (0.573%)
maximum number of reductions reached, breaking from loop
positioning took 0.9 minutes
done positioning surface
Iteration 2 =========================================
n_averages=1, current_sigma=0.5
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6407
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6429, nmarked2=0, nripped=6429
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 0.5;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=117922
Gdiag_no=-1
VMPeak = 1820472
vno start=0, stop=117922
#SI# sigma=0.5 had to be increased for 151 vertices, nripped=6429
mean border=87.9, 524 (422) missing vertices, mean dist -0.1 [0.1 (%82.8)->0.1 (%17.2))]
%91 local maxima, % 3 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0376 min
Finding expansion regions
mean absolute distance = 0.13 +- 0.19
815 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.5, host=jupyt, nav=1, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=495268.7, rms=3.691
rms = 3.7061/3.6908, sse=509985.3/495268.7, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
029: dt: 0.2500, sse=339823.2, rms=2.555 (30.772%)
030: dt: 0.2500, sse=265604.5, rms=1.756 (31.273%)
031: dt: 0.2500, sse=254916.8, rms=1.646 (6.291%)
032: dt: 0.2500, sse=244652.4, rms=1.578 (4.086%)
rms = 1.5492/1.5783, sse=246299.6/244652.4, time step reduction 2 of 3 to 0.125 0 1 1
033: dt: 0.2500, sse=246299.6, rms=1.549 (1.843%)
rms = 1.4772/1.5492, sse=256396.0/246299.6, time step reduction 3 of 3 to 0.062 0 1 0
034: dt: 0.1250, sse=256396.0, rms=1.477 (4.649%)
maximum number of reductions reached, breaking from loop
positioning took 0.5 minutes
done positioning surface
Iteration 3 =========================================
n_averages=0, current_sigma=0.25
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6429
removing 2 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6439, nmarked2=0, nripped=6439
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 0.25;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=117922
Gdiag_no=-1
VMPeak = 1928040
vno start=0, stop=117922
#SI# sigma=0.25 had to be increased for 110 vertices, nripped=6439
mean border=88.3, 565 (418) missing vertices, mean dist -0.0 [0.1 (%55.3)->0.1 (%44.7))]
%92 local maxima, % 2 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0238 min
Finding expansion regions
mean absolute distance = 0.09 +- 0.13
1636 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.2, host=jupyt, nav=0, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=254630.3, rms=1.463
rms = 1.9100/1.4635, sse=279765.3/254630.3, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
035: dt: 0.2500, sse=227375.6, rms=1.229 (16.034%)
rms = 1.1900/1.2288, sse=224595.0/227375.6, time step reduction 2 of 3 to 0.125 0 0 1
036: dt: 0.2500, sse=224595.0, rms=1.190 (3.158%)
rms = 1.1740/1.1900, sse=228522.3/224595.0, time step reduction 3 of 3 to 0.062 0 1 1
037: dt: 0.1250, sse=228522.3, rms=1.174 (1.340%)
maximum number of reductions reached, breaking from loop
positioning took 0.3 minutes
done positioning surface
Removing intersections
#ET# mris_place_surface 3.51 minutes
Writing output to ../surf/rh.white.preaparc
#VMPC# mris_place_surfaces VmPeak 1964816
mris_place_surface done
8.0.0-beta
8.0.0-beta
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical/TestSubject/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/data/structural/recon-all-clinical
mris_place_surface --adgws-in ../surf/autodet.gw.stats.rh.dat --wm wm.mgz --threads 2 --invol brain.mgz --rh --i ../surf/rh.orig --o ../surf/rh.white.preaparc --white --seg aseg.presurf.mgz --nsmooth 5
Reading in input surface ../surf/rh.orig
Smoothing surface before ripping with 5 iterations
Area 235840 0.26931 0.06148 0.013425 0.5851
Corner 707520 60.00000 9.13941 20.029026 133.8117
Edge 353760 0.79227 0.11109 0.146651 1.2577
Hinge 353760 6.38664 6.11601 0.000011 116.3464
Not reading in aparc
Reading in input volume brain.mgz
Reading in seg volume aseg.presurf.mgz
Reading in wm volume wm.mgz
MRIclipBrightWM(): nthresh=0, wmmin=5, clip=110
MRIfindBrightNonWM(): 0 bright non-wm voxels segmented.
Masking bright non-wm for white surface
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6242, nmarked2=0, nripped=6242
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247
MRISripSegs(): -2 2 0.5 ripped 0
opt weights int=0.2 loc=0 rep=5 nspr=0.3 curv=1
vertex 58961: xyz = (34.8115,3.75511,51.0703) oxyz = (34.8115,3.75511,51.0703) wxzy = (34.8115,3.75511,51.0703) pxyz = (0,0,0)
CBVO Creating mask 117922
n_averages 4
Iteration 0 =========================================
n_averages=4, current_sigma=2
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6242
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6242, nmarked2=0, nripped=6242
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 2;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=117922
Gdiag_no=-1
VMPeak = 887184
vno start=0, stop=117922
#SI# sigma=2 had to be increased for 306 vertices, nripped=6242
mean border=78.3, 502 (502) missing vertices, mean dist 0.4 [0.7 (%33.3)->0.9 (%66.7))]
%63 local maxima, %31 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.1011 min
Finding expansion regions
mean absolute distance = 0.82 +- 0.96
3399 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=2.0, host=jupyt, nav=4, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height 0.000000
momentum 0.000000
nbhd_size 0
max_nbrs 0
niterations 100
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 1
randomSeed 0
000: dt: 0.0000, sse=5354776.0, rms=15.332
001: dt: 0.5000, sse=3289522.5, rms=11.928 (22.197%)
002: dt: 0.5000, sse=2077289.5, rms=9.370 (21.449%)
003: dt: 0.5000, sse=1382639.5, rms=7.516 (19.790%)
004: dt: 0.5000, sse=988276.1, rms=6.223 (17.204%)
005: dt: 0.5000, sse=765210.3, rms=5.353 (13.980%)
006: dt: 0.5000, sse=635152.6, rms=4.768 (10.925%)
007: dt: 0.5000, sse=563391.1, rms=4.392 (7.876%)
008: dt: 0.5000, sse=515540.0, rms=4.145 (5.642%)
009: dt: 0.5000, sse=492023.0, rms=4.009 (3.279%)
010: dt: 0.5000, sse=478022.6, rms=3.906 (2.553%)
rms = 3.8606/3.9063, sse=469828.4/478022.6, time step reduction 1 of 3 to 0.250 0 0 1
011: dt: 0.5000, sse=469828.4, rms=3.861 (1.170%)
012: dt: 0.2500, sse=287934.5, rms=2.564 (33.594%)
013: dt: 0.2500, sse=248640.5, rms=2.194 (14.425%)
014: dt: 0.2500, sse=238514.9, rms=2.081 (5.127%)
015: dt: 0.2500, sse=230140.0, rms=1.996 (4.124%)
rms = 1.9594/1.9956, sse=228880.3/230140.0, time step reduction 2 of 3 to 0.125 0 0 1
016: dt: 0.2500, sse=228880.3, rms=1.959 (1.810%)
017: dt: 0.1250, sse=227234.5, rms=1.904 (2.854%)
rms = 1.8880/1.9035, sse=221167.4/227234.5, time step reduction 3 of 3 to 0.062 0 0 1
018: dt: 0.1250, sse=221167.4, rms=1.888 (0.817%)
maximum number of reductions reached, breaking from loop
positioning took 1.5 minutes
done positioning surface
Iteration 1 =========================================
n_averages=2, current_sigma=1
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6242
removing 4 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6407, nmarked2=0, nripped=6407
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 1;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=117922
Gdiag_no=-1
VMPeak = 1670604
vno start=0, stop=117922
#SI# sigma=1 had to be increased for 193 vertices, nripped=6407
mean border=85.1, 506 (426) missing vertices, mean dist -0.3 [0.4 (%90.8)->0.2 (%9.2))]
%84 local maxima, %11 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0649 min
Finding expansion regions
mean absolute distance = 0.35 +- 0.38
1352 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=1.0, host=jupyt, nav=2, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=1730947.5, rms=8.339
019: dt: 0.5000, sse=807793.8, rms=5.262 (36.902%)
020: dt: 0.5000, sse=636081.7, rms=4.467 (15.109%)
021: dt: 0.5000, sse=612913.2, rms=4.374 (2.067%)
rms = 4.4378/4.3744, sse=626413.6/612913.2, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
022: dt: 0.2500, sse=369782.8, rms=2.871 (34.374%)
023: dt: 0.2500, sse=296359.0, rms=2.153 (25.008%)
024: dt: 0.2500, sse=277288.0, rms=1.927 (10.493%)
025: dt: 0.2500, sse=267265.1, rms=1.851 (3.959%)
rms = 1.8076/1.8506, sse=265935.3/267265.1, time step reduction 2 of 3 to 0.125 0 0 1
026: dt: 0.2500, sse=265935.3, rms=1.808 (2.327%)
027: dt: 0.1250, sse=259442.5, rms=1.728 (4.393%)
rms = 1.7183/1.7282, sse=263697.9/259442.5, time step reduction 3 of 3 to 0.062 0 1 1
028: dt: 0.1250, sse=263697.9, rms=1.718 (0.573%)
maximum number of reductions reached, breaking from loop
positioning took 0.9 minutes
done positioning surface
Iteration 2 =========================================
n_averages=1, current_sigma=0.5
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6407
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6429, nmarked2=0, nripped=6429
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 0.5;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=117922
Gdiag_no=-1
VMPeak = 1820476
vno start=0, stop=117922
#SI# sigma=0.5 had to be increased for 151 vertices, nripped=6429
mean border=87.9, 524 (422) missing vertices, mean dist -0.1 [0.1 (%82.8)->0.1 (%17.2))]
%91 local maxima, % 3 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0371 min
Finding expansion regions
mean absolute distance = 0.13 +- 0.19
815 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.5, host=jupyt, nav=1, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=495268.7, rms=3.691
rms = 3.7061/3.6908, sse=509985.3/495268.7, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
029: dt: 0.2500, sse=339823.2, rms=2.555 (30.772%)
030: dt: 0.2500, sse=265604.5, rms=1.756 (31.273%)
031: dt: 0.2500, sse=254916.8, rms=1.646 (6.291%)
032: dt: 0.2500, sse=244652.4, rms=1.578 (4.086%)
rms = 1.5492/1.5783, sse=246299.6/244652.4, time step reduction 2 of 3 to 0.125 0 1 1
033: dt: 0.2500, sse=246299.6, rms=1.549 (1.843%)
rms = 1.4772/1.5492, sse=256396.0/246299.6, time step reduction 3 of 3 to 0.062 0 1 0
034: dt: 0.1250, sse=256396.0, rms=1.477 (4.649%)
maximum number of reductions reached, breaking from loop
positioning took 0.5 minutes
done positioning surface
Iteration 3 =========================================
n_averages=0, current_sigma=0.25
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6429
removing 2 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6439, nmarked2=0, nripped=6439
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 118.5830460;
border_low = 69.0000000;
outside_low = 58.4975280;
outside_hi = 118.5830460;
sigma = 0.25;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0.5
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=117922
Gdiag_no=-1
VMPeak = 1928044
vno start=0, stop=117922
#SI# sigma=0.25 had to be increased for 110 vertices, nripped=6439
mean border=88.3, 565 (418) missing vertices, mean dist -0.0 [0.1 (%55.3)->0.1 (%44.7))]
%92 local maxima, % 2 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0240 min
Finding expansion regions
mean absolute distance = 0.09 +- 0.13
1636 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning surface subiter 0
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.2, host=jupyt, nav=0, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=254630.3, rms=1.463
rms = 1.9100/1.4635, sse=279765.3/254630.3, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
035: dt: 0.2500, sse=227375.6, rms=1.229 (16.034%)
rms = 1.1900/1.2288, sse=224595.0/227375.6, time step reduction 2 of 3 to 0.125 0 0 1
036: dt: 0.2500, sse=224595.0, rms=1.190 (3.158%)
rms = 1.1740/1.1900, sse=228522.3/224595.0, time step reduction 3 of 3 to 0.062 0 1 1
037: dt: 0.1250, sse=228522.3, rms=1.174 (1.340%)
maximum number of reductions reached, breaking from loop
positioning took 0.3 minutes
done positioning surface
Removing intersections
#ET# mris_place_surface 3.56 minutes
Writing output to ../surf/rh.white.preaparc
#VMPC# mris_place_surfaces VmPeak 1964824
mris_place_surface done
Generating cortex label... RemoveHipAmgy=0
NucAccIsMedialWall=0
mris->useRealRAS=0
3 non-cortical segments detected
only using segment with 6187 vertices
erasing segment 1 (vno[0] = 62796)
erasing segment 2 (vno[0] = 71062)
Generating cortex label... RemoveHipAmgy=1
NucAccIsMedialWall=0
mris->useRealRAS=0
4 non-cortical segments detected
only using segment with 5411 vertices
erasing segment 1 (vno[0] = 60860)
erasing segment 2 (vno[0] = 62796)
erasing segment 3 (vno[0] = 71062)
Generating cortex label... RemoveHipAmgy=0
NucAccIsMedialWall=0
mris->useRealRAS=0
5 non-cortical segments detected
only using segment with 6059 vertices
erasing segment 1 (vno[0] = 65281)
erasing segment 2 (vno[0] = 70107)
erasing segment 3 (vno[0] = 70281)
erasing segment 4 (vno[0] = 70300)
Generating cortex label... RemoveHipAmgy=1
NucAccIsMedialWall=0
mris->useRealRAS=0
5 non-cortical segments detected
only using segment with 5285 vertices
erasing segment 1 (vno[0] = 38117)
erasing segment 2 (vno[0] = 39555)
erasing segment 3 (vno[0] = 65281)
erasing segment 4 (vno[0] = 70107)
smoothing for 3 iterations
setting seed for random number generator to 1234
smoothing surface tessellation for 3 iterations...
smoothing complete - recomputing first and second fundamental forms...
smoothing for 3 iterations
setting seed for random number generator to 1234
smoothing surface tessellation for 3 iterations...
smoothing complete - recomputing first and second fundamental forms...
Reading ./lh.smoothwm
avg radius = 45.9 mm, total surface area = 74964 mm^2
step 060: RMS=0.019 (target=0.015) writing inflated surface to ./lh.inflated
writing sulcal depths to ./lh.sulc
inflation complete.
inflation took 0.4 minutes
mris_inflate utimesec 24.712154
mris_inflate stimesec 0.132023
mris_inflate ru_maxrss 185780
mris_inflate ru_ixrss 0
mris_inflate ru_idrss 0
mris_inflate ru_isrss 0
mris_inflate ru_minflt 49031
mris_inflate ru_majflt 18
mris_inflate ru_nswap 0
mris_inflate ru_inblock 4392
mris_inflate ru_oublock 9368
mris_inflate ru_msgsnd 0
mris_inflate ru_msgrcv 0
mris_inflate ru_nsignals 0
mris_inflate ru_nvcsw 72
mris_inflate ru_nivcsw 99
Reading ./rh.smoothwm
avg radius = 46.7 mm, total surface area = 73946 mm^2
step 060: RMS=0.018 (target=0.015) writing inflated surface to ./rh.inflated
writing sulcal depths to ./rh.sulc
inflation complete.
inflation took 0.4 minutes
mris_inflate utimesec 24.511248
mris_inflate stimesec 0.135929
mris_inflate ru_maxrss 182836
mris_inflate ru_ixrss 0
mris_inflate ru_idrss 0
mris_inflate ru_isrss 0
mris_inflate ru_minflt 48273
mris_inflate ru_majflt 0
mris_inflate ru_nswap 0
mris_inflate ru_inblock 0
mris_inflate ru_oublock 9224
mris_inflate ru_msgsnd 0
mris_inflate ru_msgrcv 0
mris_inflate ru_nsignals 0
mris_inflate ru_nvcsw 27
mris_inflate ru_nivcsw 633
setting seed for random number generator to 1234
total integrated curvature = 27.546*4pi (346.157) --> -27 handles
ICI = 122.8, FI = 1192.5, variation=18642.116
writing Gaussian curvature to ./lh.white.preaparc.K...
done.
writing mean curvature to ./lh.white.preaparc.H...
lh.white.preaparc.H
mris_curvature done.
setting seed for random number generator to 1234
normalizing curvature values.
averaging curvature patterns 5 times.
sampling 10 neighbors out to a distance of 10 mm
202 vertices thresholded to be in k1 ~ [-0.18 0.31], k2 ~ [-0.10 0.05]
total integrated curvature = 0.674*4pi (8.474) --> 0 handles
ICI = 1.4, FI = 7.1, variation=126.493
113 vertices thresholded to be in [-0.00 0.01]
writing Gaussian curvature to ./lh.inflated.K...
thresholding curvature at 99.90% level
curvature mean = 0.000, std = 0.001
155 vertices thresholded to be in [-0.11 0.16]
done.
writing mean curvature to ./lh.inflated.H...
curvature mean = -0.016, std = 0.020
lh.inflated.H
mris_curvature done.
setting seed for random number generator to 1234
total integrated curvature = 31.097*4pi (390.773) --> -30 handles
ICI = 124.2, FI = 1199.3, variation=18725.480
writing Gaussian curvature to ./rh.white.preaparc.K...
done.
writing mean curvature to ./rh.white.preaparc.H...
rh.white.preaparc.H
mris_curvature done.
setting seed for random number generator to 1234
normalizing curvature values.
averaging curvature patterns 5 times.
sampling 10 neighbors out to a distance of 10 mm
231 vertices thresholded to be in k1 ~ [-0.20 0.32], k2 ~ [-0.08 0.06]
total integrated curvature = 0.708*4pi (8.894) --> 0 handles
ICI = 1.4, FI = 7.6, variation=132.220
119 vertices thresholded to be in [-0.00 0.01]
writing Gaussian curvature to ./rh.inflated.K...
thresholding curvature at 99.90% level
curvature mean = 0.000, std = 0.001
143 vertices thresholded to be in [-0.13 0.16]
done.
writing mean curvature to ./rh.inflated.H...
curvature mean = -0.017, std = 0.021
rh.inflated.H
mris_curvature done.
setting seed for random number genererator to 1234
version: 8.0.0-beta
available threads: 1
reading original vertex positions...
projecting onto sphere...
surface projected - minimizing metric distortion...
scaling brain by 0.307...
MRISunfold() max_passes = 1 -------
tol=5.0e-01, sigma=0.0, host=jupyt, nav=1024, nbrs=2, l_area=1.000, l_dist=1.000
using quadratic fit line minimization
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 1.000000
desired_rms_height -1.000000
momentum 0.900000
nbhd_size 7
max_nbrs 8
niterations 25
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 0
randomSeed 1234
🎨 Viewing aseg.mgz
Now we will open the aseg.mgz file. This is FreeSurfer’s automatic brain segmentation, where different brain structures are given unique numbers and colors.
- 📂
aseg.mgz= FreeSurfer’s anatomical segmentation file - 🎨 Each color = a different brain structure (e.g. hippocampus, ventricles, cortex)
- 🧩 Helps us check if FreeSurfer labelled the brain correctly
- ➕ Crosshairs again line up the same spot across all views
➡️ Click the cell below, then press Command + Enter (Mac) or Control + Enter (Windows/Linux).
This will open aseg.mgz in the viewer with FreeSurfer colors.
# Display aseg.mgz in FreeSurfer colors, labels 0–100
import tempfile, os
from pathlib import Path
ASEG_PATH = "./data/structural/recon-all-clinical/TestSubject/mri/aseg.mgz"
assert Path(ASEG_PATH).exists(), f"Missing aseg: {ASEG_PATH}"
aseg_img = nib.load(ASEG_PATH)
aseg = np.rint(aseg_img.get_fdata()).astype(np.int32)
# Keep only labels 0–100, everything else to 0
aseg = np.where((aseg >= 0) & (aseg <= 100), aseg, 0)
# Save temporary integer NIfTI
tmp_path = os.path.join(tempfile.gettempdir(), "aseg_fs_0to100.nii.gz")
nib.save(nib.Nifti1Image(aseg, aseg_img.affine, aseg_img.header), tmp_path)
# Viewer
nv = NiiVue(
height=600,
multiplanar_layout="GRID",
multiplanar_show_render=True,
is_colorbar=True,
is_orient_cube=True,
is_radiological_convention=True,
back_color=(0.1, 0.1, 0.1, 1.0),
)
# Use FreeSurfer colormap
nv.add_volume({
"path": tmp_path,
"name": "aseg",
"opacity": 1.0,
"is_label": True,
"colormap": "freesurfer",
})
nv.set_crosshair_color([0,0,0,1])
display(nv)
Image(url='https://raw.githubusercontent.com/neurodesk/example-notebooks/refs/heads/main/books/images/recon_all_clinical_aseg.png')
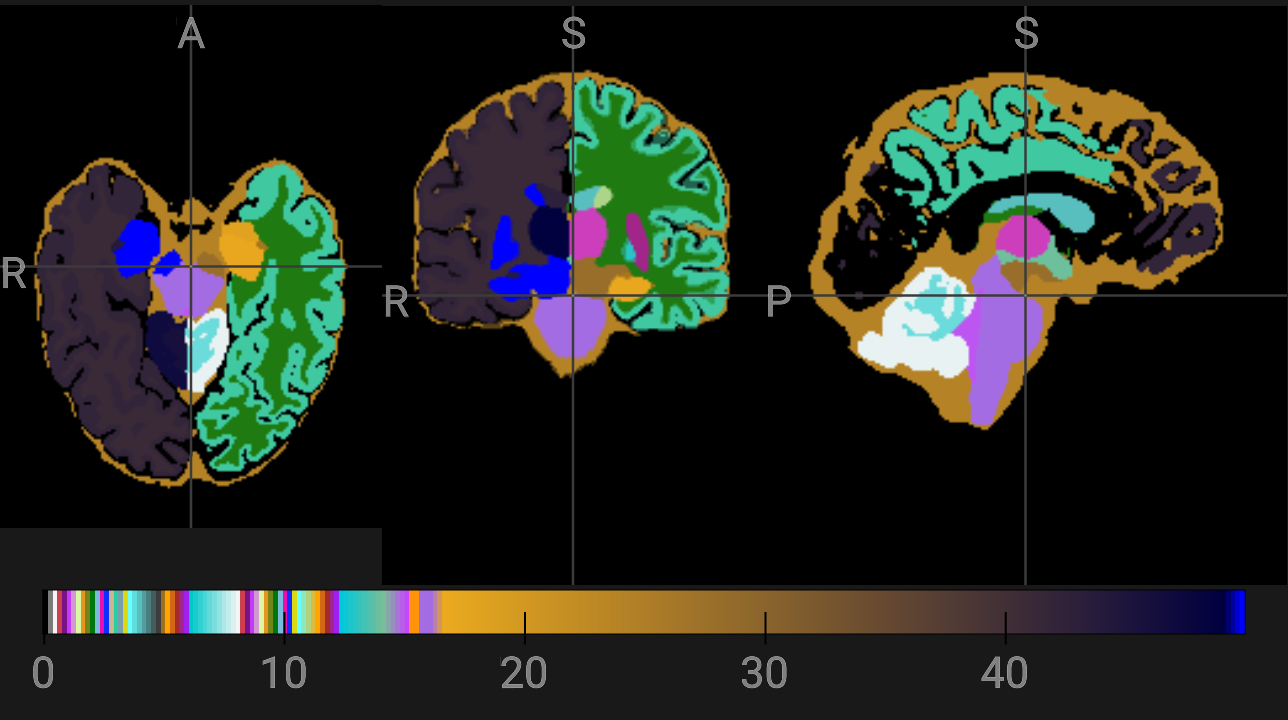
Viewing the Pial Surfaces
Now we will look at the pial surfaces. These are '3D' models of the outer surface of the brain (the grey matter boundary). FreeSurfer creates one for each hemisphere.
- 📂
lh.pial= left hemisphere surface - 📂
rh.pial= right hemisphere surface - 🔴 Left hemisphere is shown in red
- 🔵 Right hemisphere is shown in blue
- Displayed in a fully interactive 3D view (you can rotate and zoom)
➡️ Click the cell below, then press Command + Enter (Mac) or Control + Enter (Windows/Linux).
You’ll see a 3D model of the brain surface – red for left, blue for right.
# Pial surfaces with explicit per-mesh color + shader (ipyniivue)
from ipyniivue import Mesh
SUBJECTS_DIR = "./data/structural/recon-all-clinical"
SUBJECT = "TestSubject"
surf_dir = Path(SUBJECTS_DIR) / SUBJECT / "surf"
lh = surf_dir / "lh.pial"
rh = surf_dir / "rh.pial"
assert lh.exists() and rh.exists(), "Missing pial surfaces"
nv = NiiVue(
height=600,
is_colorbar=False,
is_orient_cube=True,
is_radiological_convention=True,
back_color=(0.05, 0.05, 0.05, 1.0),
)
# Use rgba255 (0..255 ints) — more reliable across builds than 0..1 floats
nv.add_mesh(Mesh(path=str(lh), name="lh.pial", rgba255=[255, 90, 90, 255], opacity=1.0))
nv.add_mesh(Mesh(path=str(rh), name="rh.pial", rgba255=[90, 90, 255, 255], opacity=1.0))
display(nv)
Image(url='https://raw.githubusercontent.com/neurodesk/example-notebooks/refs/heads/main/books/images/recon_all_clinical_pial_surface.png')
⚪ Viewing the White Matter Surfaces
Now we will open the white matter surfaces. These are 3D models of the inner boundary of the cortex, where the grey matter meets the white matter. They are used by FreeSurfer to measure cortical thickness.
- 📂
lh.white= left hemisphere white surface - 📂
rh.white= right hemisphere white surface - 🟠 Left hemisphere is shown in orange
- 🔵 Right hemisphere is shown in light blue
- 🌐 Fully interactive 3D display – rotate and zoom to explore
➡️ Click the cell below, then press Command + Enter (Mac) or Control + Enter (Windows/Linux).
You’ll see the white matter surfaces in 3D (orange = left, blue = right).
# White surfaces (lh/rh) with solid colors
lh = surf_dir/"lh.white"; rh = surf_dir/"rh.white"
assert lh.exists() and rh.exists(), "Missing white surfaces"
nv = NiiVue(height=600,is_colorbar=False, is_orient_cube=True,
is_radiological_convention=True, back_color=(0.05,0.05,0.05,1.0))
nv.add_mesh(Mesh(path=str(lh), name="lh.white", rgba255=[255,200,90,255], opacity=1.0))
nv.add_mesh(Mesh(path=str(rh), name="rh.white", rgba255=[90,200,255,255], opacity=1.0))
display(nv)
Image(url='https://raw.githubusercontent.com/neurodesk/example-notebooks/refs/heads/main/books/images/recon_all_clinical_white_matter.png')
🌐 Viewing the Inflated Surface
Now we will look at the inflated brain surface. This is a special 3D model where the folds (sulci and gyri) are “smoothed out” so the entire cortex can be seen more clearly. It is very useful for visualising activity maps or large-scale anatomy without folds hiding important areas.
- 📂
lh.inflated= inflated left hemisphere surface - 🔴 Left hemisphere is shown here in light red
- 👁️ The sulci (valleys) are expanded so you can see regions that are normally hidden inside folds
- 🌐 Fully interactive 3D view – rotate and zoom to explore the cortical sheet
➡️ Click the cell below, then press Command + Enter (Mac) or Control + Enter (Windows/Linux).
You’ll see the inflated left hemisphere in 3D.
# Inflated surfaces (lh/rh) with solid colors
lh = surf_dir/"lh.inflated"; rh = surf_dir/"rh.inflated"
assert lh.exists() and rh.exists(), "Missing inflated surfaces"
nv = NiiVue(height=600, is_colorbar=False, is_orient_cube=True,
is_radiological_convention=True, back_color=(0.05,0.05,0.05,1.0))
nv.add_mesh(Mesh(path=str(lh), name="lh.inflated", rgba255=[255,140,140,255], opacity=1.0))
#nv.add_mesh(Mesh(path=str(rh), name="rh.inflated", rgba255=[140,140,255,255], opacity=1.0))
display(nv)
Image(url='https://raw.githubusercontent.com/neurodesk/example-notebooks/refs/heads/main/books/images/recon_all_clinical_inflated.png')
🗂️ Inspecting the Subject Folder & Logs
This step shows the top-level contents of the subject’s FreeSurfer folder and the run logs. Use it to confirm that key outputs exist and to find logs if something went wrong.
SUBJECTS_DIR = ./data/structural/recon-all-clinical
SUBJECT = TestSubject
Path = ./data/structural/recon-all-clinical/TestSubject
📁 What the main folders mean
| Folder | Purpose |
|---|---|
mri/ |
All volumetric outputs (.mgz), e.g. native.mgz, aseg.mgz |
surf/ |
Cortical meshes (.white, .pial, .inflated) and annotations (.annot) |
label/ |
Region labels and color tables (.ctab) |
stats/ |
Per-region thickness/surface area stats (for group analysis) |
scripts/ |
Run logs, IsRunning flags, and command histories |
scripts/ for recent log files and any IsRunning* flags.
➡️ Click the cell below, then press Command + Enter (Mac) or Control + Enter (Windows/Linux) to list the directory tree and show log files.
# Show directory layout and recon logs
import os, subprocess, textwrap
SUBJECTS_DIR = "./data/structural/recon-all-clinical"
SUBJECT = "TestSubject"
base = f"{SUBJECTS_DIR}/{SUBJECT}"
print("=== Subject directory tree (top) ===")
subprocess.run(["bash","-lc", f'ls -lh {base}'])
print("\n=== Logs/scripts ===")
scripts = f"{base}/scripts"
subprocess.run(["bash","-lc", f'ls -lh {scripts} 2>/dev/null || true'])
notes = [
("mri/", "All volumetric outputs (.mgz)."),
("surf/", "Cortical meshes and annotation files (.white, .pial, .inflated, .annot)."),
("label/", "Labels, annotation tables (.ctab), region lists."),
("stats/", "Cortex thickness/surface area stats per region (for group analysis)."),
("scripts/", "Run logs, IsRunning flags, and command histories."),
]
for d, t in notes:
print(f"{d:12s} {t}")
=== Subject directory tree (top) ===
total 32K
drwxr-sr-x 2 jovyan users 4.0K Oct 30 06:24 label
drwxr-sr-x 3 jovyan users 4.0K Oct 30 22:18 mri
drwxr-sr-x 2 jovyan users 4.0K Oct 30 23:53 scripts
drwxr-sr-x 2 jovyan users 4.0K Oct 30 06:24 stats
drwxr-sr-x 2 jovyan users 4.0K Oct 30 23:15 surf
drwxr-sr-x 2 jovyan users 4.0K Oct 30 04:30 tmp
drwxr-sr-x 2 jovyan users 4.0K Oct 30 04:30 touch
drwxr-sr-x 2 jovyan users 4.0K Oct 30 04:30 trash
=== Logs/scripts ===
total 352K
-rw-r--r-- 1 jovyan users 347K Oct 30 23:53 recon-all-clinical.log
mri/ All volumetric outputs (.mgz).
surf/ Cortical meshes and annotation files (.white, .pial, .inflated, .annot).
label/ Labels, annotation tables (.ctab), region lists.
stats/ Cortex thickness/surface area stats per region (for group analysis).
scripts/ Run logs, IsRunning flags, and command histories.
Dependencies in Jupyter/Python#
Using the package watermark to document system environment and software versions used in this notebook
%load_ext watermark
%watermark
%watermark --iversions
Last updated: 2025-10-31T00:04:58.447546+00:00
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.16.1
Compiler : GCC 12.3.0
OS : Linux
Release : 5.4.0-204-generic
Machine : x86_64
Processor : x86_64
CPU cores : 32
Architecture: 64bit
IPython : 8.16.1
numpy : 2.2.6
nibabel : 5.2.1
ipyniivue: 2.3.2

 thomshaw92
thomshaw92 0000-0003-2490-0532
0000-0003-2490-0532